Joss-reviews: [REVIEW]: CRBHits: Conditional Reciprocal Best Hits in R
Submitting author: @kullrich (Kristian Ullrich)
Repository: https://gitlab.gwdg.de/mpievolbio-it/crbhits
Version: 0.0.2
Editor: @KristinaRiemer
Reviewer: @clauswilke, @a-r-j
Archive: 10.5281/zenodo.4247942
:warning: JOSS reduced service mode :warning:
Due to the challenges of the COVID-19 pandemic, JOSS is currently operating in a "reduced service mode". You can read more about what that means in our blog post.
Status
Status badge code:
HTML: <a href="https://joss.theoj.org/papers/26d66e8db73048560958b65c1714588c"><img src="https://joss.theoj.org/papers/26d66e8db73048560958b65c1714588c/status.svg"></a>
Markdown: [](https://joss.theoj.org/papers/26d66e8db73048560958b65c1714588c)
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@clauswilke & @a-r-j, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
- Make sure you're logged in to your GitHub account
- Be sure to accept the invite at this URL: https://github.com/openjournals/joss-reviews/invitations
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @KristinaRiemer know.
✨ Please try and complete your review in the next six weeks ✨
Review checklist for @clauswilke
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@kullrich) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
Review checklist for @a-r-j
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@kullrich) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
All 73 comments
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @clauswilke, @a-r-j it looks like you're currently assigned to review this paper :tada:.
:warning: JOSS reduced service mode :warning:
Due to the challenges of the COVID-19 pandemic, JOSS is currently operating in a "reduced service mode". You can read more about what that means in our blog post.
:star: Important :star:
If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿
To fix this do the following two things:
- Set yourself as 'Not watching' https://github.com/openjournals/joss-reviews:

- You may also like to change your default settings for this watching repositories in your GitHub profile here: https://github.com/settings/notifications

For a list of things I can do to help you, just type:
@whedon commands
For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type:
@whedon generate pdf
Reference check summary:
OK DOIs
- 10.1371/journal.pgen.1004365 is OK
- doi:10.21105/joss.00142 is OK
- 10.1101/gr.113985.110 is OK
- 10.1186/s13059-015-0721-2 is OK
- 10.1093/protein/12.2.85 is OK
- 10.1007/978-3-540-35306-5_10 is OK
- 10.1016/S1672-0229(10)60008-3 is OK
- 10.1007/BF02407308 is OK
- 10.18129/B9.bioc.Biostrings is OK
- 10.1093/oxfordjournals.molbev.a026236 is OK
- 10.1016/j.cbpa.2003.12.004 is OK
- 10.1093/nar/gkr853 is OK
- 10.1007/978-1-4939-9074-0_5 is OK
- 10.1016/S0022-2836(05)80360-2 is OK
- 10.1371/journal.pgen.1000304 is OK
- 10.1016/0022-2836(81)90087-5 is OK
MISSING DOIs
- None
INVALID DOIs
- None
@a-r-j I think you need to accept the invite to review in order to be listed as an assignee here. Then both reviewers can feel free to start your review! The checklists above and reviewer guidelines are good places to start. Let me know if you have questions or need help.
Hiya, I've completed my review. Nice work, @kullrich. The only thing that appears to be missing are some contribution guidelines. I see you have a section on bug reporting; could someone (@KristinaRiemer ?) advise whether this is sufficient for JOSS.
I similarly couldn't find any community guidelines. In addition, I think both the high-level documentation of the package and the paper could be improved somewhat. The first sentence in the paper states "CRBHits is a reimplementation of the
Conditional Reciprocal Best Hit (CRBH) algorithm crb-blast in R." The paper then continues discussing the history of the CRBH algorithm, which is fine. However, the package also has additional features, such as aligning sequences and calculating evolutionary rates, and these topics don't entirely fit into the framework that this is just a "reimplementation of CRBH". I think you need to reconsider the framing, so that users are not surprised to find other features in the package.
Similarly, the vignettes need some work. All three vignettes start with installation instructions, which is confusing. I would expect to find this material only in the basic vignette. Also, the basic vignette should give a brief overview over all different functionality of the package, so the user can understand how everything fits together. All vignette sections are extremely terse and difficult to follow for somebody who isn't intimately familiar with the specific material already. A little more explanatory text would help. In many places, the vignette also looks more like notes than a finished document (see e.g. Section 3 in the dN/dS clustering vignette; I also note the change in notation here: dN/dS or Ka/Ks?). Finally, the "paranome" vignette is entirely empty, so I suggest you either write it or disable it for now.
Submission of the package to CRAN or Bioconductor would be good, but is not required for publication.
Hi, I am currently on summer vacation with my family and will be back in two weeks.
I will work on the two unfinished vignettes and will work on the high-level documentation as mentioned by @clauswilke.
I will move the installation section to be fully visible in the basic vignette and will only set links in the other two.
I will also rethink the framing how to include the downstream functionality more clearly in the title. I will definitely not remove these downstream functionalities for this package even if users would be hopefully positively surprised finding them in this package.
However, since calculating dN/dS values (or kA/kS values, some authors use one notation for comparing only within one species and other use it for between species nucleotide diversity calculation) is a wide field I am not sure how to sufficiently address this in this paper more deeply.
I could also try to submit the package to CRAN or Bioconductor, however, since the external dependencies mainly LAST is only available for unix systems (no windows executables) it might be difficult to submit to CRAN or Bioconductor (both check for windows functionality). I could create an option to switch to original BLAST executables for windows systems (makes the sequence search slow again) or use an alternative like DIAMOND. But I need to think about this if it is worth spending time on the windows issue, since within win10 there is an easy option to install a virtual Ubuntu on a windows machine from the win10 store for free.
I actually have not tried Cygwin yet but will try on a windows machine if I can build LAST and directly use it on win10 which is checked with the travis.yml in the end (here to be able to create the vignettes for a proper R check, all dependencies are built in a virtual environment and need to compile before the R package is installed). If you have experience with this external dependency windows C++ Cygwin issue, it would be great to get some hints how to efficiently solve it.
Best regards
Kristian
--
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Genetics
Max Planck Institute for Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected]
--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.
On 22. Jul 2020, at 20:45, Claus Wilke notifications@github.com wrote:
I similarly couldn't find any community guidelines. In addition, I think both the high-level documentation of the package and the paper could be improved somewhat. The first sentence in the paper states "CRBHits is a reimplementation of the
Conditional Reciprocal Best Hit (CRBH) algorithm crb-blast in R." The paper then continues discussing the history of the CRBH algorithm, which is fine. However, the package also has additional features, such as aligning sequences and calculating evolutionary rates, and these topics don't entirely fit into the framework that this is just a "reimplementation of CRBH". I think you need to reconsider the framing, so that users are not surprised to find other features in the package.Similarly, the vignettes need some work. All three vignettes start with installation instructions, which is confusing. I would expect to find this material only in the basic vignette. Also, the basic vignette should give a brief overview over all different functionality of the package, so the user can understand how everything fits together. All vignette sections are extremely terse and difficult to follow for somebody who isn't intimately familiar with the specific material already. A little more explanatory text would help. In many places, the vignette also looks more like notes than a finished document (see e.g. Section 3 in the dN/dS clustering vignette; I also note the change in notation here: dN/dS or Ka/Ks?). Finally, the "paranome" vignette is entirely empty, so I suggest you either write it or disable it for now.
Submission of the package to CRAN or Bioconductor would be good, but is not required for publication.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub https://github.com/openjournals/joss-reviews/issues/2424#issuecomment-662621967, or unsubscribe https://github.com/notifications/unsubscribe-auth/AB5ZWI7FGODMWRLAZJEARL3R44XTFANCNFSM4OPEJNXA.
Hi,
regarding the community guidelines to contribute to the package, I have seen so far this options for JOSS R package papers and I will think about one solution for CRBHits.
Best regards
Kristian
e.g.: https://gitlab.inria.fr/miet/miet/-/tree/master https://gitlab.inria.fr/miet/miet/-/tree/master
Which rely on https://www.contributor-covenant.org/ https://www.contributor-covenant.org/
or https://github.com/PhilBoileau/scPCA
https://github.com/PhilBoileau/scPCA/blob/master/CONTRIBUTING.md https://github.com/PhilBoileau/scPCA/blob/master/CONTRIBUTING.md
or https://github.com/Sage-Bionetworks/mhealthtools
Contributing and Reporting Issues
To report an issue, please file a GitHub issue with the main repo https://github.com/Sage-Bionetworks/mHealthTools.
If you would like to contribute to mhealthtools, please file an issue so that we can establish a statement of need, avoid redundant work, and track progress on your contribution. Once an issue has been filed and we've identified how to best orient your contribution with package development as a whole, fork http://help.github.com/fork-a-repo/ the main repo https://github.com/Sage-Bionetworks/mHealthTools, branch off a feature branch https://www.google.com/search?q=git+feature+branches from develop, commit http://git-scm.com/docs/git-commit and push http://git-scm.com/docs/git-push your changes to your fork and submit a pull request http://help.github.com/send-pull-requests/ for Sage-Bionetworks/mhealthtools:develop.
--
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Genetics
Max Planck Institute for Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected]
--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.
On 24. Jul 2020, at 22:00, Kristian Ullrich ullrich@evolbio.mpg.de wrote:
Hi, I am currently on summer vacation with my family and will be back in two weeks.
I will work on the two unfinished vignettes and will work on the high-level documentation as mentioned by @clauswilke.
I will move the installation section to be fully visible in the basic vignette and will only set links in the other two.
I will also rethink the framing how to include the downstream functionality more clearly in the title. I will definitely not remove these downstream functionalities for this package even if users would be hopefully positively surprised finding them in this package.
However, since calculating dN/dS values (or kA/kS values, some authors use one notation for comparing only within one species and other use it for between species nucleotide diversity calculation) is a wide field I am not sure how to sufficiently address this in this paper more deeply.
I could also try to submit the package to CRAN or Bioconductor, however, since the external dependencies mainly LAST is only available for unix systems (no windows executables) it might be difficult to submit to CRAN or Bioconductor (both check for windows functionality). I could create an option to switch to original BLAST executables for windows systems (makes the sequence search slow again) or use an alternative like DIAMOND. But I need to think about this if it is worth spending time on the windows issue, since within win10 there is an easy option to install a virtual Ubuntu on a windows machine from the win10 store for free.
I actually have not tried Cygwin yet but will try on a windows machine if I can build LAST and directly use it on win10 which is checked with the travis.yml in the end (here to be able to create the vignettes for a proper R check, all dependencies are built in a virtual environment and need to compile before the R package is installed). If you have experience with this external dependency windows C++ Cygwin issue, it would be great to get some hints how to efficiently solve it.
Best regards
Kristian
--
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Genetics
Max Planck Institute for Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected] ullrich@evolbio.mpg.de--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.On 22. Jul 2020, at 20:45, Claus Wilke <[email protected] notifications@github.com> wrote:
I similarly couldn't find any community guidelines. In addition, I think both the high-level documentation of the package and the paper could be improved somewhat. The first sentence in the paper states "CRBHits is a reimplementation of the
Conditional Reciprocal Best Hit (CRBH) algorithm crb-blast in R." The paper then continues discussing the history of the CRBH algorithm, which is fine. However, the package also has additional features, such as aligning sequences and calculating evolutionary rates, and these topics don't entirely fit into the framework that this is just a "reimplementation of CRBH". I think you need to reconsider the framing, so that users are not surprised to find other features in the package.Similarly, the vignettes need some work. All three vignettes start with installation instructions, which is confusing. I would expect to find this material only in the basic vignette. Also, the basic vignette should give a brief overview over all different functionality of the package, so the user can understand how everything fits together. All vignette sections are extremely terse and difficult to follow for somebody who isn't intimately familiar with the specific material already. A little more explanatory text would help. In many places, the vignette also looks more like notes than a finished document (see e.g. Section 3 in the dN/dS clustering vignette; I also note the change in notation here: dN/dS or Ka/Ks?). Finally, the "paranome" vignette is entirely empty, so I suggest you either write it or disable it for now.
Submission of the package to CRAN or Bioconductor would be good, but is not required for publication.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub https://github.com/openjournals/joss-reviews/issues/2424#issuecomment-662621967, or unsubscribe https://github.com/notifications/unsubscribe-auth/AB5ZWI7FGODMWRLAZJEARL3R44XTFANCNFSM4OPEJNXA.
I agree with both of the reviewers that the community guidelines could use some improvement. Adding a CONTRIBUTING such as the one you linked to above @kullrich would be useful. Do note that contribution guidelines are separate, though complementary, to a code of conduct like the Contributor Covenant.
@KristinaRiemer I have now added the Code of Conduct and Contributing section. Maybe you can check if this is sufficient for JOSS.
Concerning submitting the package to CRAN or Bioconductor using Cygwin I was able to build the external resources on win10.
However it is still a bit complicated to parse the automatic R check (Cygwin is not included in travis.ci, while MSYS is, but MSYS uses the windows memory allocation headers, the external resource is using the unix headers like Cygwin).
In short words, without re-writing a part of the external resources to stick to the windows memory allocation, it might be hard to get a working windows binary according to CRAN or Bioconductor rules. Another issue would be to distribute the win10 binaries, since you need to include the DLLs of the building system (either Cygwin or MSYS) so that the binaries work out of the box for the windows user. I already contacted the developer of the external resource LAST and he said that in his opinion it would be fine under the license conditions of Cygwin and LAST. I will keep you posted on that issue.
Best regards
Kristian
I think the contribution guidelines and code of conduct are sufficient, and want to make sure the reviewers @a-r-j and @clauswilke agree?
@kullrich I think you may want to revisit @clauswilke's suggestions about the documentation. Specifically either filling out or removing the empty paranome vignette and the third section of the dNdS vignette. The first line of the second section of the dNdS vignette could also use more prose about the purpose of that section, too. Lastly, including the additional functionalities beyond the algorithm reimplementation in the first paragraph of the paper would be useful. Let me know if you have other questions about this.
Also, you do not need to submit this to CRAN or Bioconductor.
@KristinaRiemer I agree with all of your points.
Yes, I am in agreement too.
@kullrich do you have any questions about these documentation/paper improvements? These should be fairly minor and straightforward to implement.
No, there was just no time due to two kids at home and COVID-19 pandemic regulations for school and kindergarden in Germany.
--
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Genetics
Max Planck Institute for Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected]
--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.
----- Ursprüngliche Mail -----
Von: "Kristina Riemer" notifications@github.com
An: "openjournals/joss-reviews" joss-reviews@noreply.github.com
CC: "Kristian Ullrich" ullrich@evolbio.mpg.de, "Mention" mention@noreply.github.com
Gesendet: Dienstag, 8. September 2020 17:21:13
Betreff: Re: [openjournals/joss-reviews] [REVIEW]: CRBHits: Conditional Reciprocal Best Hits in R (#2424)
@kullrich do you have any questions about these documentation/paper improvements? These should be fairly minor and straightforward to implement.
--
You are receiving this because you were mentioned.
Reply to this email directly or view it on GitHub:
https://github.com/openjournals/joss-reviews/issues/2424#issuecomment-688950192
@kullrich it's all good, just wanted to check in. These are especially difficult times.
Hi @kullrich, how's it going? Do you have an idea of when you'll have a chance to address these changes?
Hi,
I think I can finish earliest in two weeks.
Best regards
Kristian
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected]
--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.
On 13. Oct 2020, at 16:24, Kristina Riemer notifications@github.com wrote:
Hi @kullrich https://github.com/kullrich, how's it going? Do you have an idea of when you'll have a chance to address these changes?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub https://github.com/openjournals/joss-reviews/issues/2424#issuecomment-707774593, or unsubscribe https://github.com/notifications/unsubscribe-auth/AB5ZWIY4GJXMCQBEOY7ZJSTSKRPKPANCNFSM4OPEJNXA.
Sounds good! Let me know if I can help in any way.
Dear Kristina Riemer,
I am almost done with restructuring and finalising the vignettes.
Due to mandatory steps in the processing of the input data I added three more functions.
- directly extract longest isoform CDS from NCBI or Ensembl URL input
- extract gene position from CDS from NCBI or Ensembl URL input
- assign tandem duplicates
With these changes it is now possible to directly get a dot plot for Ka/Ks values between two species
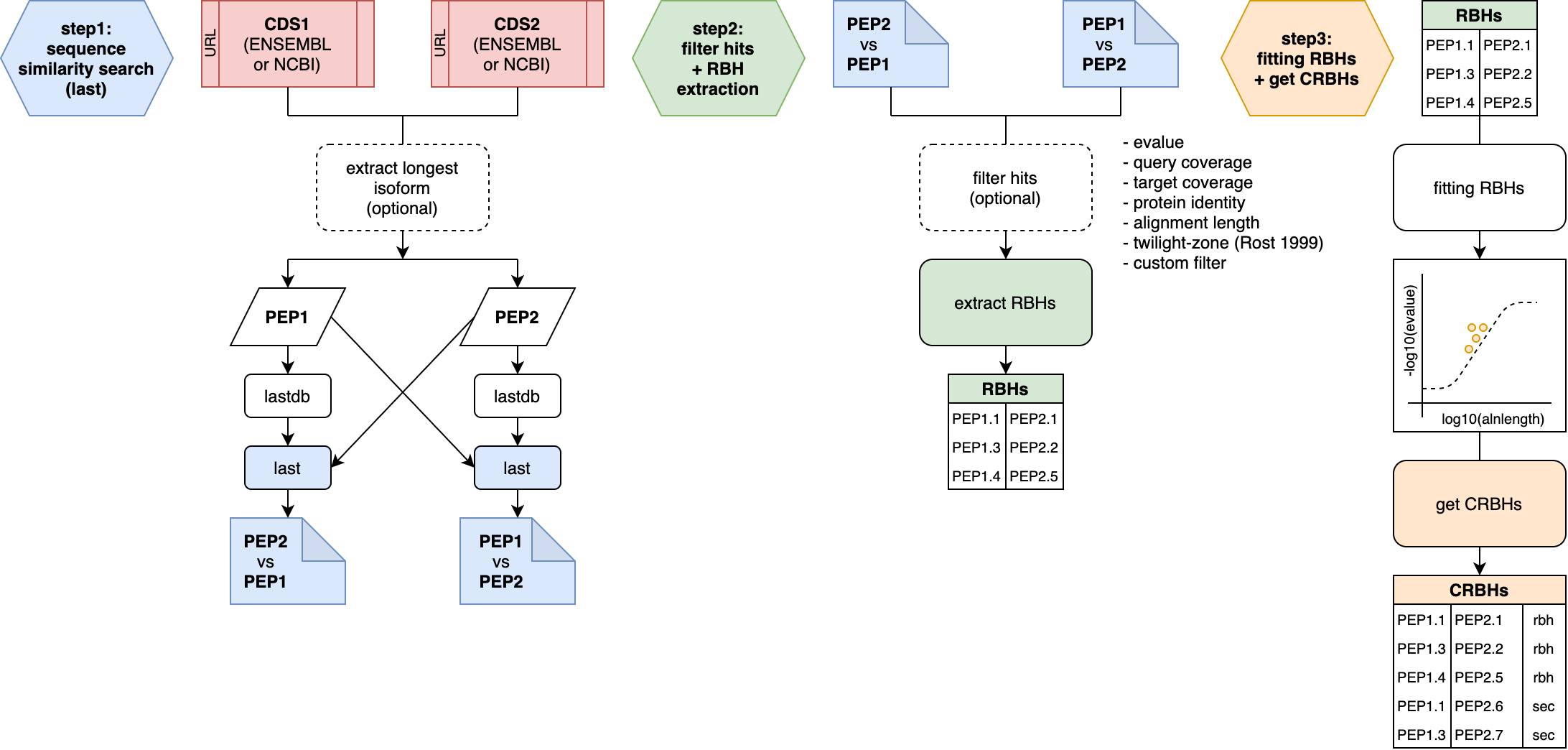
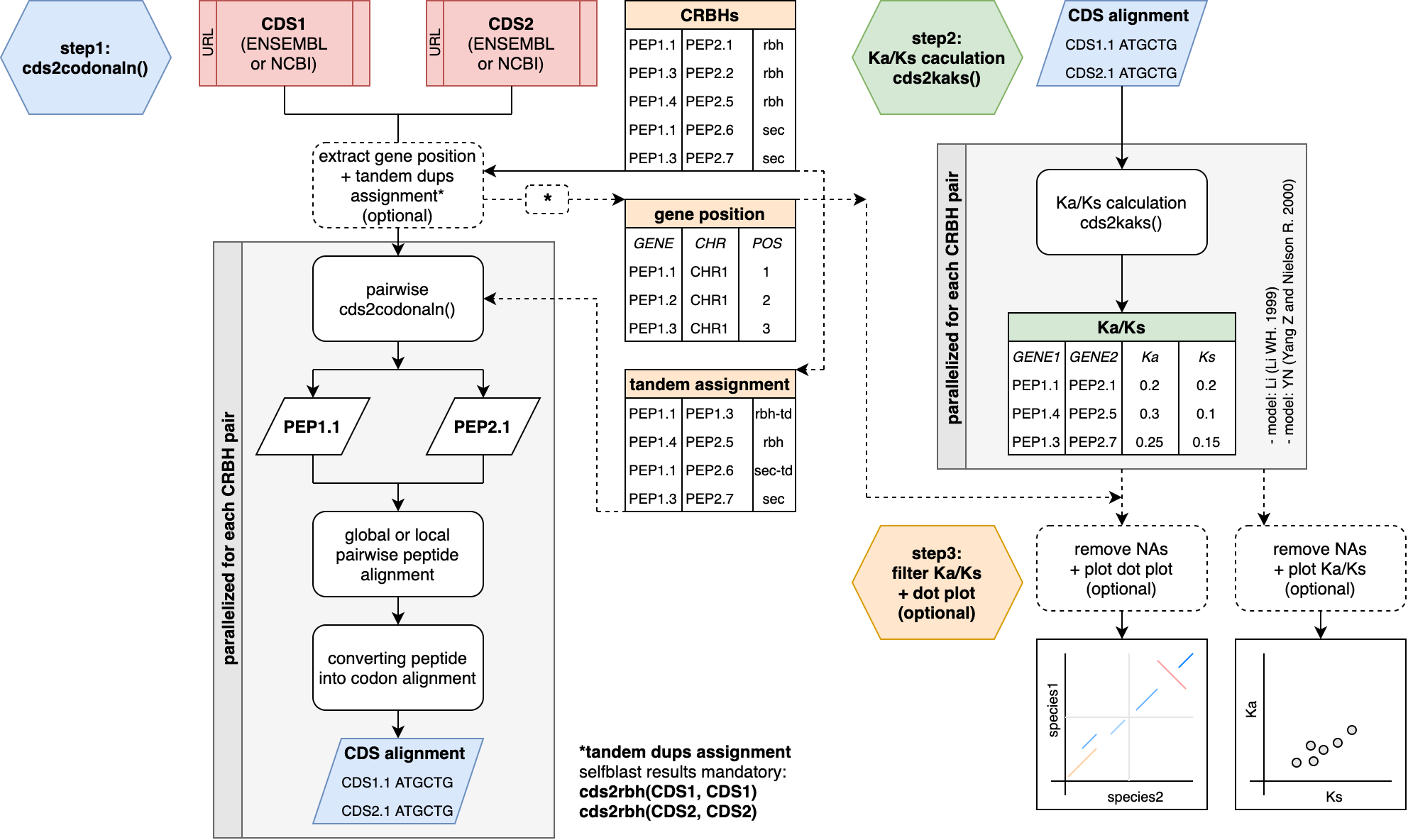
However, I also now added two workflow images that describe the tow main high-order functions (cds2rbh; rbh2kaks) of the package in more detail.
The question is if one of these should go into the paper as a figure?
Best regards and thank you in anticipation
Kristian Ullrich
--
Kristian Ullrich, Ph.D.
Scientific IT group
Department of Evolutionary Biology
August Thienemann Str. 2
24306 Plön
Germany
+49 4522 763 313
[email protected]
--
CONFIDENTIALITY NOTICE:
The contents of this email message and any attachments are intended solely for the addressee(s) and may contain confidential and/or privileged information and may be legally protected from disclosure. If you are not the intended recipient, you are hereby notified that any use, dissemination, copying, or storage of this message or its attachments is strictly prohibited.
On 13. Oct 2020, at 17:05, Kristina Riemer notifications@github.com wrote:
Sounds good! Let me know if I can help in any way.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub https://github.com/openjournals/joss-reviews/issues/2424#issuecomment-707803981, or unsubscribe https://github.com/notifications/unsubscribe-auth/AB5ZWI7CGNM6HLNEDFTI473SKRUDZANCNFSM4OPEJNXA.
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
Hi @KristinaRiemer,
I was now able to finish the paper, it has the following changes as suggested by @clauswilke.
The Title is changed into: "CRBHits: From Conditional Reciprocal Best Hits to Codon Alignments and Ka/Ks in R"
to better directly address the wider framework of the R package.
In addition, the summary now includes in the first two sentences:
"CRBHits is a coding sequence (CDS) analysis pipeline in R. It reimplements the Conditional Reciprocal Best Hit (CRBH) algorithm and covers all necessary steps from sequence similarity searches, codon alignments to Ka/Ks calculations and synteny."
Figure1 now covers the two main functions and its main steps from CRBHit pairs to Ka/Ks values.
In the Basic Vignette both function are explained in more detail by its own figures, which I will attach to this post.
Example one was changed since now it is possible and most likely quite useful to directly parse the input URL for coding sequences to extract the "longest isoform" from NCBI or ENSEMBL.
I have changed the section "Downstream functionalities" into "Coding sequence analysis and synteny"
In the same section I have added the importance and short background of gene duplication classification via DAGchainer, which is now also part of CRBHits.
All external tools are forked within CRBHits and aggree with the MIT licence. They can be easily compiled with "make.last()", "make.KaKs_Calculator2()" and "make.dagchainer()".
To demonstrate this functionality, a second example was added highlighting a selfblast of the model plant Arabidopsis thaliana.
The figure 3 shows the results of the Ka/Ks analysis and the synteny analysis by cross-referencing each other data by the color scheme, as noted in the figure caption as follows.
"Selfblast CRBHit pair results for Arabidopsis thaliana. (A) DAGchainer dotplot per chromosome colored by syntenic group and (B) colored by Ks. (C) Histogram of Ks values colored by syntenic group."
There was a runtime decrease, since I changed to a quicker CDS to amino acid conversion.
I hope to finish the last part of the KaKs Vignette tomorrow and give the repo a new status flag.
Finally, as suggessted by @clauswilke, the Whole-Genome-Duplication prediction vignette was removed from this version.
Best regards
Kristian
I'm fine with the revised manuscript. No more comments from my side.
I'm happy too! Nice work 👍
@whedon check references
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1371/journal.pgen.1004365 is OK
- doi:10.21105/joss.00142 is OK
- 10.1101/gr.113985.110 is OK
- 10.1186/s13059-015-0721-2 is OK
- 10.1093/protein/12.2.85 is OK
- 10.1007/978-3-540-35306-5_10 is OK
- 10.1016/S1672-0229(10)60008-3 is OK
- 10.1007/BF02407308 is OK
- 10.18129/B9.bioc.Biostrings is OK
- 10.1093/oxfordjournals.molbev.a026236 is OK
- 10.1016/j.cbpa.2003.12.004 is OK
- 10.1093/nar/gkr853 is OK
- 10.1007/978-1-4939-9074-0_5 is OK
- 10.1016/S0022-2836(05)80360-2 is OK
- 10.1371/journal.pgen.1000304 is OK
- 10.1016/0022-2836(81)90087-5 is OK
- 10.1093/bioinformatics/bth397 is OK
MISSING DOIs
- None
INVALID DOIs
- https://doi.org/10.1007/978-3-642-86659-3 is INVALID because of 'https://doi.org/' prefix
Great, thanks @kullrich for making all of those improvements. And thank you for your reviews @a-r-j & @clauswilke!
@kullrich in reviewing the article's references, I found a couple of typos that should be fixed:
- The Smith & Waterman 1981 reference has an "& others" in the author string that can be removed, as it looks like there are only the two authors for that publication
- As shown in the whedon references check above, the "https://doi.org/" part of the Ohno 1970 DOI should be removed
- Most of the journal titles need to have capitalizations, e.g., "Journal of molecular biology" should be "Journal of Molecular Biology"
Once you are finished with those improvements, the next steps are to: 1) create a tagged release on GitHub and 2) archive the code (on Zenodo, figshare, institutional repository, etc.) to generate a DOI. If you can post a link to the DOI here when you're done please.
@whedon generate pdf
10.5281/zenodo.4247942
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
Hi @KristinaRiemer,
as you suggested the repo v0.0.2 is now hosted on zenodo.
The repo on github was now also set to be a public repo like on gitlab.
Best regards
Kristian
@a-r-j I checked the "community guidelines" part of your review, you said earlier you were satisfied with it but let me know if that's not the case.
@whedon check references
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1371/journal.pgen.1004365 is OK
- doi:10.21105/joss.00142 is OK
- 10.1101/gr.113985.110 is OK
- 10.1186/s13059-015-0721-2 is OK
- 10.1093/protein/12.2.85 is OK
- 10.1007/978-3-540-35306-5_10 is OK
- 10.1016/S1672-0229(10)60008-3 is OK
- 10.1007/BF02407308 is OK
- 10.18129/B9.bioc.Biostrings is OK
- 10.1093/oxfordjournals.molbev.a026236 is OK
- 10.1016/j.cbpa.2003.12.004 is OK
- 10.1093/nar/gkr853 is OK
- 10.1007/978-1-4939-9074-0_5 is OK
- 10.1016/S0022-2836(05)80360-2 is OK
- 10.1371/journal.pgen.1000304 is OK
- 10.1016/0022-2836(81)90087-5 is OK
- 10.1093/bioinformatics/bth397 is OK
- 10.1007/978-3-642-86659-3 is OK
MISSING DOIs
- None
INVALID DOIs
- None
@whedon set 10.5281/zenodo.4247942 as archive
OK. 10.5281/zenodo.4247942 is the archive.
@whedon set 0.0.2 as version
OK. 0.0.2 is the version.
@whedon accept
Attempting dry run of processing paper acceptance...
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1371/journal.pgen.1004365 is OK
- doi:10.21105/joss.00142 is OK
- 10.1101/gr.113985.110 is OK
- 10.1186/s13059-015-0721-2 is OK
- 10.1093/protein/12.2.85 is OK
- 10.1007/978-3-540-35306-5_10 is OK
- 10.1016/S1672-0229(10)60008-3 is OK
- 10.1007/BF02407308 is OK
- 10.18129/B9.bioc.Biostrings is OK
- 10.1093/oxfordjournals.molbev.a026236 is OK
- 10.1016/j.cbpa.2003.12.004 is OK
- 10.1093/nar/gkr853 is OK
- 10.1007/978-1-4939-9074-0_5 is OK
- 10.1016/S0022-2836(05)80360-2 is OK
- 10.1371/journal.pgen.1000304 is OK
- 10.1016/0022-2836(81)90087-5 is OK
- 10.1093/bioinformatics/bth397 is OK
- 10.1007/978-3-642-86659-3 is OK
MISSING DOIs
- None
INVALID DOIs
- None
:wave: @openjournals/joss-eics, this paper is ready to be accepted and published.
Check final proof :point_right: https://github.com/openjournals/joss-papers/pull/1908
If the paper PDF and Crossref deposit XML look good in https://github.com/openjournals/joss-papers/pull/1908, then you can now move forward with accepting the submission by compiling again with the flag deposit=true e.g.
@whedon accept deposit=true
I'm sorry @kullrich, I'm afraid I can't do that. That's something only editor-in-chiefs are allowed to do.
Hi @kullrich! Things are looking good. Just one comment on your paper and then we are done! Looking through your references, I see at least a couple items in titles that are not capitalized but should be: R and Python. Please go through your references carefully and add {} around words or the full titles in which you want to preserve capitalization.
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
Hi @kthyng, do I need to move the flag v0.0.2 of the release to the current version and update the zenodo file deposition with the new release file?
@kthyng checked and changed the paper.bib file accordingly to preserve capitalization, also carefully re-checked and updated the runtimes in table1. Since I have also put the dependencies on bioconda, I updated the README for the whole package, which should not have any consequences for the manuscript.
Hi @kthyng, do I need to move the flag v0.0.2 of the release to the current version and update the zenodo file deposition with the new release file?
This isn't super important since they are minor changes.
@whedon accept deposit=true
Doing it live! Attempting automated processing of paper acceptance...
🐦🐦🐦 👉 Tweet for this paper 👈 🐦🐦🐦
🚨🚨🚨 THIS IS NOT A DRILL, YOU HAVE JUST ACCEPTED A PAPER INTO JOSS! 🚨🚨🚨
Here's what you must now do:
- Check final PDF and Crossref metadata that was deposited :point_right: https://github.com/openjournals/joss-papers/pull/1911
- Wait a couple of minutes to verify that the paper DOI resolves https://doi.org/10.21105/joss.02424
- If everything looks good, then close this review issue.
Party like you just published a paper! 🎉🌈🦄💃👻🤘
Any issues? Notify your editorial technical team...
Congrats on your new publication @kullrich! Thanks to editor @KristinaRiemer and reviewers @clauswilke and @a-r-j for your time and expertise!! 🎉
(I'll close this issue once the DOI resolves)
:tada::tada::tada: Congratulations on your paper acceptance! :tada::tada::tada:
If you would like to include a link to your paper from your README use the following code snippets:
Markdown:
[](https://doi.org/10.21105/joss.02424)
HTML:
<a style="border-width:0" href="https://doi.org/10.21105/joss.02424">
<img src="https://joss.theoj.org/papers/10.21105/joss.02424/status.svg" alt="DOI badge" >
</a>
reStructuredText:
.. image:: https://joss.theoj.org/papers/10.21105/joss.02424/status.svg
:target: https://doi.org/10.21105/joss.02424
This is how it will look in your documentation:
We need your help!
Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
- Volunteering to review for us sometime in the future. You can add your name to the reviewer list here: https://joss.theoj.org/reviewer-signup.html
- Making a small donation to support our running costs here: https://numfocus.org/donate-to-joss
Congrats @kullrich and thanks to @a-r-j & @clauswilke for your work!
@KristinaRiemer & @a-r-j & @clauswilke thank you for handling this and for all the input made.
I am happy that the R package is now published and hope that it will be frequently used in future.
Congrats @kullrich ! Nice work :)
Most helpful comment
@a-r-j I think you need to accept the invite to review in order to be listed as an assignee here. Then both reviewers can feel free to start your review! The checklists above and reviewer guidelines are good places to start. Let me know if you have questions or need help.