Rdkit: Issue with inversion/retention of stereochemistry
- RDKit Version: 2019.09.3.0
- OS: Mac
- Are you using conda? Yes
- If you are using conda, which channel did you install the rdkit from? rdkit
Description:
Ran into a few strange behaviors (at least relative to what I would expect) when trying to code up a substitution reaction. Minimal examples reproduced below:
For all examples the reactant of interest is Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
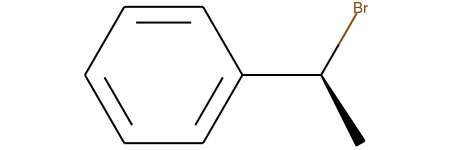
We begin by testing the simple bimolecular substitution reaction, with inversion of stereochemistry at the reaction center:
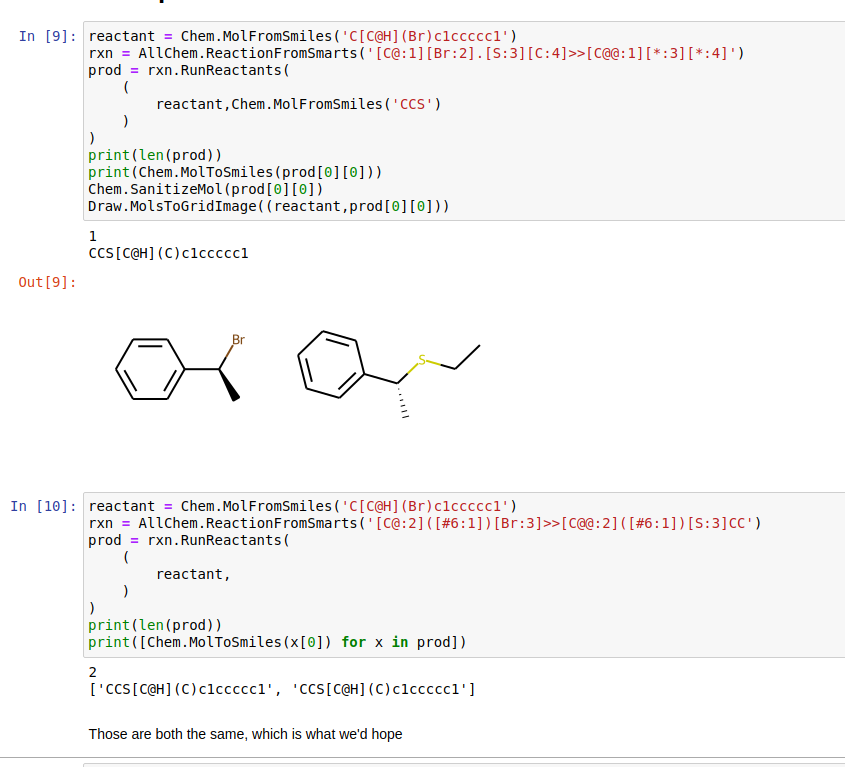
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:1][Br:2].[S:3][C:4]>>[C@@:1][*:3][*:4]')
prod = rxn.RunReactants(
(
reactant,Chem.MolFromSmiles('CCS')
)
)
print(len(prod))
print(Chem.MolToSmiles(prod[0][0]))
The result is 'CCS[C@@H](C)c1ccccc1' (shown below)
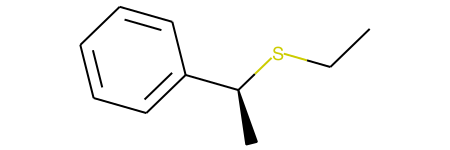
However, this is actually not the expected inverted product. The reason for this I believe is due to the change of the order of the SMILES. The carbon indeed changes from [C@] to [C@@]; however, since the canonicalization changes the bond we look down to define clockwise/counter-clockwise, no true inversion occurs. Ultimately, I did not see an easy way to fix this problem (though I welcome suggestions!), so started testing a unimolecular transformation, as shown below.
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([#6:1])[Br:3]>>[C@@:2]([#6:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This example, produces both stereoisomers:
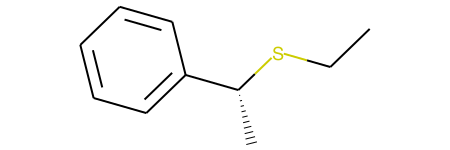
and
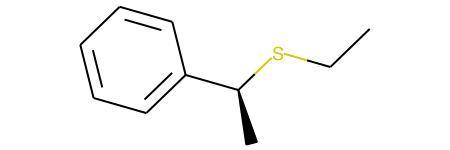
I understand that my RXN SMARTS is likely overly general, since '[C@:2]([#6:1])[Br:3]' may be matched in two different ways to the reactant. However, either way it matches, shouldn't the product simply invert the stereochemistry of the original match? It is a bit unclear to me why the configuration of the reactant is retained.
Investigating this behavior, I tried the less general rxn below
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([C:1])[Br:3]>>[C@@:2]([C:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This actually only produces the expected product with inversion of stereochemistry:
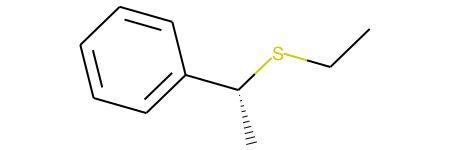
Lastly, I tried to isolate the behavior of the aromatic carbon causing troubles:
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([c:1])[Br:3]>>[C@@:2]([c:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This reaction only produces the unexpected product with retention.
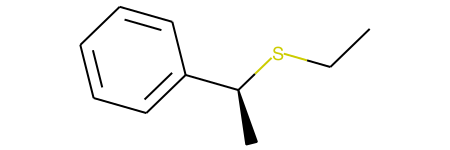
I expect that an issue may arise because the original SMILES of the reactant, ''C[C@H](Br)c1ccccc1' is defining counter-clockwise with respect to looking down the non-aromatic CC bond, and not the cC bond.
However, I am pretty confused in general, so would really appreciate any clarification. I apologize for the lengthy issue, but hopefully the reproducible examples are of help.
All 3 comments
Thanks for that detailed and well described bug report!
There's definitely something odd/unexpected going on here that I'm going to have to dig into (it's not a trivial topic) in order to understand/explain.
@mc-robinson : I believe we have this fixed on master now. It'll be in the 2020.09.1 release (coming in mid-October).
Here are the new results for a couple of your examples:
There's a gist with more here:
https://gist.github.com/greglandrum/6f5912c1ab5b29610c8c49ca62bf75a2
Thanks @greglandrum!
Most helpful comment
@mc-robinson : I believe we have this fixed on master now. It'll be in the 2020.09.1 release (coming in mid-October).

Here are the new results for a couple of your examples:
There's a gist with more here:
https://gist.github.com/greglandrum/6f5912c1ab5b29610c8c49ca62bf75a2