Keras: Concurrent training of two models with shared weights
Hello,
I would like to train a network where two datasets with different features map to the same set of classes. Idea is to have both inputs each have a separate ReLU layer first which will then converge on another, common ReLU layer which will then be sent to the Softmax. The two tasks are different but very related so I hope the synergistic training of the final layer will generate better results for each dataset compared to training them separately.
At this point the only way I can think of to do this is to manually setup the minibatches and run for loops where I update the first model on one batch using .train_on_batch, copy the weights of the last two layers (the second ReLU and the Softmax) to the other model and then update the second model, and so on... Is there a simpler way using the Keras Functional API? And if not, can you consider this as a feature request?
As a note, I know how to setup multi-input networks using .concatenate but such networks seem to consider both data points coming from different branches as a single example. In addition, I have reasons against simply merging the datasets and training them all together on a simple feedforward network.
I would greatly appreciate any help, thank you very much...
All 4 comments
I believe that copying the weights is not necessary if you reuse the layers (in opposite of just the weights) when creating the second model. Both models will share the reference to the same layer and, therefore, react to any changes made by each others training processes:
from keras import Model, Input
from keras.layers import Dense
shape = [28 * 28]
units = 256
classes = 32
x, y = Input(shape=shape), Input(shape=shape)
shared = Dense(units, activation='relu', name='shared')
predictions = Dense(classes, activation='softmax', name='predictions')
zx = predictions(shared(Dense(units, activation='relu', name='x_limb')(x)))
zy = predictions(shared(Dense(units, activation='relu', name='y_limb')(y)))
model_x = Model(inputs=[x], outputs=[zx])
model_y = Model(inputs=[y], outputs=[zy])
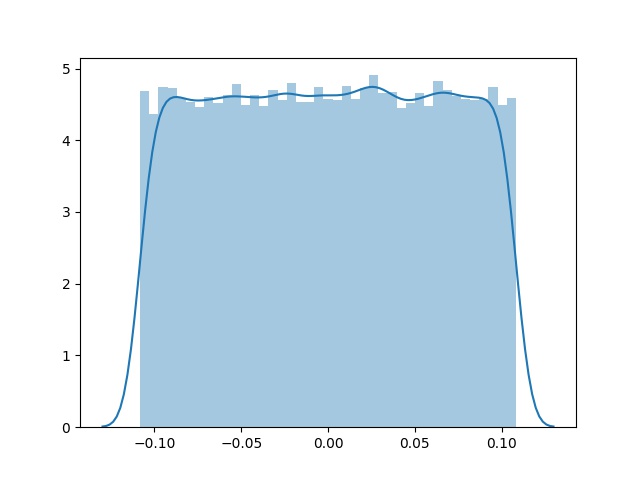
You can see model_y training indeed changes model_x weights by plotting their distribution before and after training:
from keras import backend as K
from keras.datasets import mnist
import matplotlib.pyplot as plt
import seaborn as sns
x_shared_weights, x_shared_bias = model_x.get_layer('shared').weights
sns.distplot(K.eval(x_shared_weights).ravel())
plt.savefig('x_shared_w_e0.jpg')
plt.clf()
# Training...
(data_tr, target_tr), (data_test, target_test) = mnist.load_data()
data_tr, data_test = (d.reshape(-1, 28 * 28) for d in (data_tr, data_test))
model_y.compile(optimizer='adam', loss='sparse_categorical_crossentropy')
model_y.fit(data_tr, target_tr, epochs=5, batch_size=32)
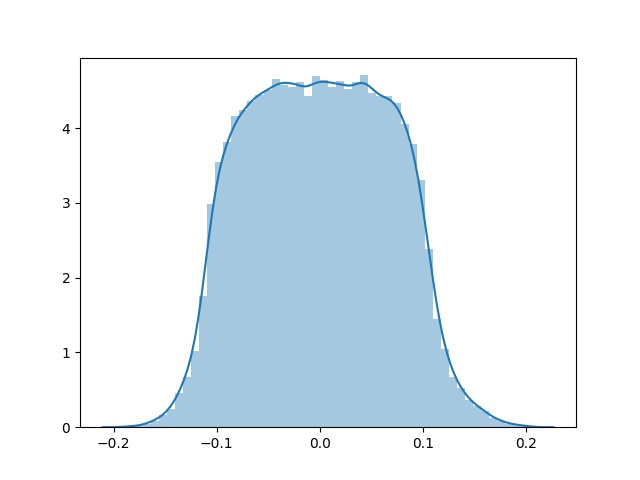
sns.distplot(K.eval(x_shared_weights).ravel())
plt.savefig('x_shared_w_e5.jpg')
plt.clf()
Before:
After 5 epochs:
Thank you very much, I have a few follow-ups if you don't mind:
So I implemented this as such:
`input1= Input(shape=(610,))
input2 = Input(shape=(642,))
x1 = layers.Dense(200, kernel_regularizer=regularizers.l2(0.002), activation='relu')(input1)
x2 = layers.Dense(200, kernel_regularizer=regularizers.l2(0.002), activation='relu')(input2 )
sharedReLU = layers.Dense(200, kernel_regularizer=regularizers.l2(0.002), activation='relu')
sharedDO = layers.Dropout(0.5)
sharedSM = layers.Dense(207, kernel_regularizer=regularizers.l2(0.002), activation='softmax')
output_tensor1 = sharedSM(sharedDO(sharedReLU(x1)))
output_tensor2 = sharedSM(sharedDO(sharedReLU(x2)))
model1 = Model(input1, output_tensor1)
model2 = Model(input2, output_tensor2)`
The first problem I'm noticing is this: it will throw an error if the sizes of the unshared dense layers need to be different or if I need to connect the shared layer directly to the different sized inputs. I suppose in those cases the only way is to copy weights. Also this got to me thinking that if it has to impose a single input shape to the shared layer, would the weights between the first and second dense layers also be shared? This is obviously completely undesirable as I would like to have those learned independently. Finally after the training is complete, I would have expected the weights of the last layers for two models be exactly the same; but I checked this manually and they don't appear to be. Am I missing something?
Besides all this, I would like to reiterate that the core of my question was actually concerning the simultaneous training of these two networks that share layers; since I'm not trying to take advantage of a pre-trained network; both need to be trained from scratch and I expect both to take advantage of each other. This is how I'm currently training (in the weight-copying case, if I used the method you suggest as above I would simply delete the .set_weights lines):
`eps = 20
for i in range(eps):
history = model1.fit(x1, y1, epochs=1+i, initial_epoch= i, batch_size=128,
callbacks=callbacks_list1, validation_split=0.1, shuffle=True,
class_weight=d_class_weights)
val_loss1.append(history.history['val_loss']); val_acc1.append(history.history['val_acc'])
loss1.append(history.history['loss']); acc1.append(history.history['acc'])
model2.layers[4].set_weights(model1.layers[4].get_weights())
history = model2.fit(x2, y2, epochs=1+i,initial_epoch= i, batch_size=128,
callbacks=callbacks_list2, validation_split=0.1, shuffle=True,
class_weight=d_class_weights)
val_loss2.append(history.history['val_loss']); val_acc2.append(history.history['val_acc'])
loss2.append(history.history['loss']); acc2.append(history.history['acc']);
model1.layers[4].set_weights(model2.layers[4].get_weights())`
There are several things that are not optimal about this. First is having to record history manually which is not a big deal. But I also noticed some callbacks that depend on history doesn't work properly, such as EarlyStopping based on val_loss (though oddly, ModelCheckpoint based on val_acc works fine). Also, ideally I would like the models to update each other after every minibatch not epoch, which is again possible through switching after every .train_on_batch instance but even more complicated to implement. I am essentially wondering if there's an easier way to train the models together?
Thank you...
The first problem I'm noticing is this: it will throw an error if the sizes of the unshared dense layers need to be different or if I need to connect the shared layer directly to the different sized inputs.
I'm not quite sure how would this work. It seems a little non-conventional, at least.
These models share layer sharedReLU. This layer's input distribution should be the same regardless where it's coming from.
I was actually imagining you had two non-structured input streams (like images). In such cases, you could:
- pad the stream with smaller perceptual field to match the larger one
- crop the larger to fit the the smaller's shape
- employ some layer that normalized the input field with respect to its size (e.g.
GlobalAveragePooling2D)
If you are dealing with two streams of semantic data (high-end variables) in which features intersect, you could impute the missing features from each stream using median values (e.g. SimpleImputer). This would sufficiently organize the data so the first shared layer wouldn't mistake a variable for another.
would the weights between the first and second dense layers also be shared?
Which are the first and second layers? Name these so I can understand.
In any case, the weights shared between these models are from layers sharedReLU and sharedSM. The weights from the first 2 layers with 200 units in your code do not share weights.
I also noticed some callbacks that depend on history doesn't work properly, such as EarlyStopping based on val_loss
I'm fairly confident these mechanisms are working correctly, as there are test cases validating them. It's hard to tell from my position, but maybe it's a noise/instability problem in your data?
You seem to have a lot of questions. You should try some of these on Stackoverflow. I believe people there will be better suited to help you.
[...] was actually concerning the simultaneous training of these two networks
As far as I can tell, the callbacks will not work for more than one model at the same time. If you want alternating training, then you need to implement something in the lines bellow yourself:
try:
for epoch in range(epochs):
for (x_batch, y_batch) in train_data:
losses = model1.fit_on_batch(x_batch, y_batch)
losses = model2.fit_on_batch(x_batch, y_batch)
losses1 = model1.evaluate(*valid_data)
losses2 = model2.evaluate(*valid_data)
# Model checkpoint if best loss so far...
# Early stop if losses are increasing...
catch KeyboardInterrupt:
# ...
Well, if you are exploring ideas, I'll just throw this suggestion: why don't you try to merge both input spaces and mask the features of a sample based on which dataset it belongs? Then, scale that signal with respect to the number of units not masked (similar to dropout). This might just create a second layer that is strong enough to activate when fed by both inputs, separately.
Furthermore, you will be able to use all keras callbacks, as you would effectively be training a single model. Something like this:
x1, y2 = np.random.randn(100000, 610), np.random.randint(10, shape=(100000))
x2, y2 = np.random.randn(100000, 642), np.random.randint(10, shape=(100000))
x1 = np.hstack((x1, np.zeros(642)) / 642
x2 = np.hstack((np.zeros(600), x2)) / 600
x, y = np.vstack((x1, x2)), np.vstack((y1, y2))
# shuffle the new dataset...
model = Sequential([
Dense(1024, activation='relu', input_shape=[1242]),
...
])
model.fit(x, y, ...)
Dear Lucas,
Thank you very much for all your responses, it has been very helpful. Particularly your idea about merging the inputs and masking based on the origin is really interesting and I'll definitely give it a try at some point. For now, my implementation of copying the weights after every epoch seems to be working for me and I'll stick with it. I was just exploring if there were any features of Keras that I wasn't aware of which could make the concurrent training of such two models even simpler but from what I gather the answer to that appears to be no. I will close the issue. Thanks again...
Most helpful comment
I believe that copying the weights is not necessary if you reuse the layers (in opposite of just the weights) when creating the second model. Both models will share the reference to the same layer and, therefore, react to any changes made by each others training processes:
You can see
model_ytraining indeed changesmodel_xweights by plotting their distribution before and after training:Before:

After 5 epochs:
