Keras: multi_gpu_model fails with timeseries data
It appears that multi_gpu_model does not adequately support time series datasets. The cause is most likely the get_slice function.
As seen in issue #11941 , when multiple GPUs are used the predicted results have similar patterns to those provided with a single GPU, but with a much lower range. Issue #11941 demonstrates this effect on a sinusoidal dataset where the predicted waveform is an attenuated version of the original when multiple GPUs are engaged. (A sinusoidal waveform was chosen for clarity). Issue #11941 has working code that illustrates the problem.


I suspect the cause for this is in the get_slice function in https://github.com/keras-team/keras/blob/master/keras/utils/multi_gpu_utils.py
The get_slice function splits the data in n segments and allocates each segment to one of n GPUs for processing. From my understanding of LSTM / GRU models, it is important to maintain an ongoing feedback loop for predictions to occur. By splitting up the data into n segments, each GPU is working on a 1/n th set of data, each GPU starts work on each segment without knowing the end state of the results of the previous segment.
In other words the weight calculations get interrupted n times over the dataset starting over from scratch each time.
The concatenation of the weights over the entire dataset is therefore affected as each GPU has only worked on a subset of data reducing the effectiveness of the weights calculation. This probably explains the great difference in the RMSE values of single versus multi GPU predictions and the attenuation in the charts.
def get_slice(data, i, parts):
shape = K.shape(data)
batch_size = shape[:1]
input_shape = shape[1:]
step = batch_size // parts
if i == parts - 1:
size = batch_size - step * i
else:
size = step
size = K.concatenate([size, input_shape], axis=0)
stride = K.concatenate([step, input_shape * 0], axis=0)
start = stride * i
return K.slice(data, start, size)
If my suspicion is true, then at a minimum the current multi_gpu_model documentation should advise against using it with timeseries data.
I'd also suggest the addition of a flag to change behavior for timeseries data. One approach to use could be to make each GPU work in parallel on the entire dataset (epoch). The parallel epoch run with the best RMSE or similar criterion would be selected for the weights to be used for the subsequent epoch. It doesn't speed up calculations though, and I'd welcome an approach that does.
My concern is that this failure generates no errors with val_loss values improving with each epoch. This lulls users into thinking that there are no issues when in fact there are.
Ubuntu 18.04.1 LTS
Keras 2.2.4
Keras-Applications 1.0.6
Keras-Preprocessing 1.0.5
tensorboard 1.12.0
tensorflow-gpu 1.12.0
All 19 comments
Hi @gabrieldemarmiesse , @palisadoes ,
First; Really good summary of this issue. I started to think a bit about what the problem with the get_slicefunction could be, splitting the data into distributed batches (over >1 GPU) should be doable since the GPU works like this...however, there might be an issue working with stateful models.
Two short questions to help us along the way:
(1) Did you try using multi_gpu_model and setting the number of GPU's to one (or this is perhaps what you have done in your first example)? Can you also test using 2 and 3 GPU's?
(2) Both the predicted training (yellow) and the predicted data (green) in the latter example seems to show the correct wavelength behavior, however, the scaling is inaccurate. Can you see if the recombination of the data is correct?
I saw one more thing;
Line 122 in the multi_gpu_utils code says Example 3 - Training models with weights merge on GPU (recommended for NV-link) , did you use NV-link? Can you see if multi_gpu_modelworks the same with SLI, NV-link or no bridge? From what I have read previously, a bridge is not needed when working with Keras/TF.
On a side note; thanks again for posting this issue. I was just on my way to buy my second water cooled 1080 ti but I'll wait with this now until this item is cleared out.
Hello again @gabrieldemarmiesse , @palisadoes ,
Sequence bucketing (https://arxiv.org/ftp/arxiv/papers/1708/1708.05604.pdf) might be a way to solve the issue with get_slice() ... what do you think about this?
# create and fit the LSTM network without cpu
serial_model = Sequential()
serial_model.add(LSTM(4, input_shape=(1, look_back)))
serial_model.add(Dense(1))
get pic:
pic
@sparkingarthur,
Thanks for this info. Could you please add the complete code (at least a bit more than the model definition would be helpful)?
@sparkingarthur,
Thanks for this info. Could you please add the complete code (at least a bit more than the model definition would be helpful)?
It's almost the same as shown in the issue, and I just comment the line:
with tf.device('/cpu:0'):
#script
"""LSTM for sinusoidal data problem with regression framing.
Based on:
https://machinelearningmastery.com/time-series-prediction-lstm-recurrent-neural-networks-python-keras/
"""
import argparse
import math
import numpy
import matplotlib.pyplot as plt
from pandas import DataFrame
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.utils import multi_gpu_model
import os
import tensorflow as tf
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error
from keras.models import load_model
os.environ['CUDA_VISIBLE_DEVICES']="0,1"
def create_dataset(dataset, look_back=1):
dataX, dataY = [], []
for i in range(len(dataset)-look_back-1):
a = dataset[i:(i+look_back), 0]
dataX.append(a)
dataY.append(dataset[i + look_back, 0])
return numpy.array(dataX), numpy.array(dataY)
gpus = 2
def main():
numpy.random.seed(7)
dataframe = DataFrame(
[0.00000, 5.99000, 11.92016, 17.73121, 23.36510, 28.76553, 33.87855,
38.65306, 43.04137, 46.99961, 50.48826, 53.47244, 55.92235, 57.81349,
59.12698, 59.84970, 59.97442, 59.49989, 58.43086, 56.77801, 54.55785,
51.79256, 48.50978, 44.74231, 40.52779, 35.90833, 30.93008, 25.64279,
20.09929, 14.35496, 8.46720, 2.49484, -3.50245, -9.46474, -15.33247,
-21.04699, -26.55123, -31.79017, -36.71147, -41.26597, -45.40815,
-49.09663, -52.29455, -54.96996, -57.09612, -58.65181, -59.62146,
-59.99540, -59.76988, -58.94716, -57.53546, -55.54888, -53.00728,
-49.93605, -46.36587, -42.33242, -37.87600, -33.04113, -27.87613,
-22.43260, -16.76493, -10.92975, -4.98536, 1.00883, 6.99295, 12.90720,
18.69248, 24.29100, 29.64680, 34.70639, 39.41920, 43.73814, 47.62007,
51.02620, 53.92249, 56.28000, 58.07518, 59.29009, 59.91260, 59.93648,
59.36149, 58.19339, 56.44383, 54.13031, 51.27593, 47.90923, 44.06383,
39.77815, 35.09503, 30.06125, 24.72711, 19.14590, 13.37339, 7.46727,
1.48653, -4.50907, -10.45961, -16.30564, -21.98875, -27.45215,
-32.64127, -37.50424, -41.99248, -46.06115, -49.66959, -52.78175,
-55.36653, -57.39810, -58.85617, -59.72618, -59.99941, -59.67316,
-58.75066, -57.24115, -55.15971, -52.52713, -49.36972, -45.71902,
-41.61151, -37.08823, -32.19438, -26.97885, -21.49376, -15.79391,
-9.93625, -3.97931, 2.01738, 7.99392, 13.89059, 19.64847, 25.21002,
30.51969, 35.52441, 40.17419, 44.42255, 48.22707, 51.54971, 54.35728,
56.62174, 58.32045, 59.43644, 59.95856, 59.88160, 59.20632, 57.93947,
56.09370, 53.68747, 50.74481, 47.29512, 43.37288, 39.01727, 34.27181,
29.18392, 23.80443, 18.18710, 12.38805, 6.46522, 0.47779, -5.51441,
-11.45151])
dataset = dataframe.values
dataset = dataset.astype('float32')
scaler = MinMaxScaler(feature_range=(0, 1))
dataset = scaler.fit_transform(dataset)
train_size = int(len(dataset) * 0.67)
test_size = len(dataset) - train_size
train, test = dataset[0:train_size, :], dataset[train_size:len(dataset), :]
look_back = 1
trainX, trainY = create_dataset(train, look_back)
testX, testY = create_dataset(test, look_back)
trainX = numpy.reshape(trainX, (trainX.shape[0], 1, trainX.shape[1]))
testX = numpy.reshape(testX, (testX.shape[0], 1, testX.shape[1]))
serial_model = Sequential()
serial_model.add(LSTM(4, input_shape=(1, look_back)))
serial_model.add(Dense(1))
if gpus == 1:
parallel_model = serial_model
else:
parallel_model = multi_gpu_model(
serial_model,
gpus=2)
parallel_model.compile(
loss='mean_squared_error', optimizer='adam')
parallel_model.fit(
trainX, trainY,
epochs=100,
batch_size=int(dataset.size * gpus / 20),
verbose=2)
serial_model.save("xxx.h5")
# make predictions ####2 gpus went wrong
serial_model = load_model("xxx.h5")
if gpus == 1:
trainPredict = serial_model.predict(trainX)
testPredict = serial_model.predict(testX)
else:
trainPredict = parallel_model.predict(trainX)
testPredict = parallel_model.predict(testX)
# invert predictions
trainPredict = scaler.inverse_transform(trainPredict)
trainY = scaler.inverse_transform([trainY])
testPredict = scaler.inverse_transform(testPredict)
testY = scaler.inverse_transform([testY])
# calculate root mean squared error
trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:, 0]))
print('Train Score: %.2f RMSE' % (trainScore))
testScore = math.sqrt(mean_squared_error(testY[0], testPredict[:, 0]))
print('Test Score: %.2f RMSE' % (testScore))
# shift train predictions for plotting
trainPredictPlot = numpy.empty_like(dataset)
trainPredictPlot[:, :] = numpy.nan
trainPredictPlot[look_back:len(trainPredict)+look_back, :] = trainPredict
# shift test predictions for plotting
testPredictPlot = numpy.empty_like(dataset)
testPredictPlot[:, :] = numpy.nan
testPredictPlot[
len(trainPredict)+(look_back*2)+1:len(dataset)-1, :] = testPredict
# plot baseline and predictions
plt.plot(scaler.inverse_transform(dataset), label='Complete Data')
plt.plot(trainPredictPlot, label='Training Data')
plt.plot(testPredictPlot, label='Prediction Data')
plt.legend(loc='upper left')
plt.title('Using {} GPUs'.format(gpus))
plt.show()
if __name__ == "__main__":
main()
Besides, my gpus:
Fri May 10 20:17:05 2019
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 390.46 Driver Version: 390.46 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
|===============================+======================+======================|
| 0 Tesla V100-PCIE... On | 00000000:04:00.0 Off | 0 |
| N/A 37C P0 37W / 250W | 15985MiB / 16160MiB | 0% Default |
+-------------------------------+----------------------+----------------------+
| 1 Tesla V100-PCIE... On | 00000000:05:00.0 Off | 0 |
| N/A 37C P0 37W / 250W | 15985MiB / 16160MiB | 0% Default |
+-------------------------------+----------------------+----------------------+
| 2 Tesla V100-PCIE... On | 00000000:08:00.0 Off | 0 |
| N/A 38C P0 39W / 250W | 15377MiB / 16160MiB | 0% Default |
+-------------------------------+----------------------+----------------------+
| 3 Tesla V100-PCIE... On | 00000000:09:00.0 Off | 0 |
| N/A 39C P0 39W / 250W | 15377MiB / 16160MiB | 0% Default |
+-------------------------------+----------------------+----------------------+
and with python2.7, keras 2.2.4, tensorflow 1.8
Python 2.7.5 (default, Oct 30 2018, 23:45:53)
[GCC 4.8.5 20150623 (Red Hat 4.8.5-36)] on linux2
Type "help", "copyright", "credits" or "license" for more information.
>>> import keras
/usr/lib64/python2.7/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.
from ._conv import register_converters as _register_converters
Using TensorFlow backend.
>>> import tensorflow as tf
>>> import keras
>>> print tf.__version__
1.8.0
>>> print keras.__version__
2.2.4
>>>
Great work, @sparkingarthur!
So you believe that the get_slice() function works as intended without compromising the sequental nature of the data? (see @palisadoes initial post regarding this flaw; "without knowing the end state of the results of the previous segment")
Was there a problem working with 2 GPUs? # make predictions ####2 gpus went wrong
Impressive; Four V100's in one system. I'm looking towards two water cooled 1080 Ti's.
@pusj
The LSTM fitting function works perfectly when you are using a single graphics card. But when you use 2 gpus, the prediction result of the model has a large deviation. There is still a bias as shown in my picture when I create and fit the LSTM network without cpu ( by commenting with tf.device('/cpu:0'):) .
In a word, the result (using 2gpus, creating model without cpu:/0) is a little better than that when using cpu to create a model ( by using with tf.device('/cpu:0'):) but still worse than that using single gpu.
@sparkingarthur ,
Well put.
I understand that the result is degraded when using > 1 GPU...however, I don't see that this has to be the case. When performing the training of the single GPU, parallelization is used (correct me if I´m wrong here )...with this said one could hope that parallelization over several GPU should not be an issue (like in the example I showed above in which sequence bucketing was used (see Figure 5)). I agree that scaling (referring to computational resource) may be influenced in a multi-GPU setup, however, it should be possible to obtain the same loss for a multi-GPU setup as for a single-GPU setup.
BTW, what did you think of sequence bucketing? Something worth putting a bit of my time on?
BTW, what did you think of sequence bucketing? Something worth putting a bit of my time on?
@pusj In my opinion, sequence bucketing is useful to speed up your RNN training.
Sequence bucketing seems to solve the issue with varying input length, however, this technique does not solve the issue with the get_slice function. @sparkingarthur , do you have a better idea how to split the data? Or do you consider this to be solved by performing all steps on the GPUs?
I saw one more thing;
Line 122 in the multi_gpu_utils code says
Example 3 - Training models with weights merge on GPU (recommended for NV-link), did you use NV-link? Can you see ifmulti_gpu_modelworks the same with SLI, NV-link or no bridge? From what I have read previously, a bridge is not needed when working with Keras/TF.On a side note; thanks again for posting this issue. I was just on my way to buy my second water cooled 1080 ti but I'll wait with this now until this item is cleared out.
No I didn't use NV link.
#script """LSTM for sinusoidal data problem with regression framing. Based on: https://machinelearningmastery.com/time-series-prediction-lstm-recurrent-neural-networks-python-keras/ """ import argparse import math import numpy import matplotlib.pyplot as plt from pandas import DataFrame from keras.models import Sequential from keras.layers import Dense from keras.layers import LSTM from keras.utils import multi_gpu_model import os import tensorflow as tf from sklearn.preprocessing import MinMaxScaler from sklearn.metrics import mean_squared_error from keras.models import load_model os.environ['CUDA_VISIBLE_DEVICES']="0,1" def create_dataset(dataset, look_back=1): dataX, dataY = [], [] for i in range(len(dataset)-look_back-1): a = dataset[i:(i+look_back), 0] dataX.append(a) dataY.append(dataset[i + look_back, 0]) return numpy.array(dataX), numpy.array(dataY) gpus = 2 def main(): numpy.random.seed(7) dataframe = DataFrame( [0.00000, 5.99000, 11.92016, 17.73121, 23.36510, 28.76553, 33.87855, 38.65306, 43.04137, 46.99961, 50.48826, 53.47244, 55.92235, 57.81349, 59.12698, 59.84970, 59.97442, 59.49989, 58.43086, 56.77801, 54.55785, 51.79256, 48.50978, 44.74231, 40.52779, 35.90833, 30.93008, 25.64279, 20.09929, 14.35496, 8.46720, 2.49484, -3.50245, -9.46474, -15.33247, -21.04699, -26.55123, -31.79017, -36.71147, -41.26597, -45.40815, -49.09663, -52.29455, -54.96996, -57.09612, -58.65181, -59.62146, -59.99540, -59.76988, -58.94716, -57.53546, -55.54888, -53.00728, -49.93605, -46.36587, -42.33242, -37.87600, -33.04113, -27.87613, -22.43260, -16.76493, -10.92975, -4.98536, 1.00883, 6.99295, 12.90720, 18.69248, 24.29100, 29.64680, 34.70639, 39.41920, 43.73814, 47.62007, 51.02620, 53.92249, 56.28000, 58.07518, 59.29009, 59.91260, 59.93648, 59.36149, 58.19339, 56.44383, 54.13031, 51.27593, 47.90923, 44.06383, 39.77815, 35.09503, 30.06125, 24.72711, 19.14590, 13.37339, 7.46727, 1.48653, -4.50907, -10.45961, -16.30564, -21.98875, -27.45215, -32.64127, -37.50424, -41.99248, -46.06115, -49.66959, -52.78175, -55.36653, -57.39810, -58.85617, -59.72618, -59.99941, -59.67316, -58.75066, -57.24115, -55.15971, -52.52713, -49.36972, -45.71902, -41.61151, -37.08823, -32.19438, -26.97885, -21.49376, -15.79391, -9.93625, -3.97931, 2.01738, 7.99392, 13.89059, 19.64847, 25.21002, 30.51969, 35.52441, 40.17419, 44.42255, 48.22707, 51.54971, 54.35728, 56.62174, 58.32045, 59.43644, 59.95856, 59.88160, 59.20632, 57.93947, 56.09370, 53.68747, 50.74481, 47.29512, 43.37288, 39.01727, 34.27181, 29.18392, 23.80443, 18.18710, 12.38805, 6.46522, 0.47779, -5.51441, -11.45151]) dataset = dataframe.values dataset = dataset.astype('float32') scaler = MinMaxScaler(feature_range=(0, 1)) dataset = scaler.fit_transform(dataset) train_size = int(len(dataset) * 0.67) test_size = len(dataset) - train_size train, test = dataset[0:train_size, :], dataset[train_size:len(dataset), :] look_back = 1 trainX, trainY = create_dataset(train, look_back) testX, testY = create_dataset(test, look_back) trainX = numpy.reshape(trainX, (trainX.shape[0], 1, trainX.shape[1])) testX = numpy.reshape(testX, (testX.shape[0], 1, testX.shape[1])) serial_model = Sequential() serial_model.add(LSTM(4, input_shape=(1, look_back))) serial_model.add(Dense(1)) if gpus == 1: parallel_model = serial_model else: parallel_model = multi_gpu_model( serial_model, gpus=2) parallel_model.compile( loss='mean_squared_error', optimizer='adam') parallel_model.fit( trainX, trainY, epochs=100, batch_size=int(dataset.size * gpus / 20), verbose=2) serial_model.save("xxx.h5") # make predictions ####2 gpus went wrong serial_model = load_model("xxx.h5") if gpus == 1: trainPredict = serial_model.predict(trainX) testPredict = serial_model.predict(testX) else: trainPredict = parallel_model.predict(trainX) testPredict = parallel_model.predict(testX) # invert predictions trainPredict = scaler.inverse_transform(trainPredict) trainY = scaler.inverse_transform([trainY]) testPredict = scaler.inverse_transform(testPredict) testY = scaler.inverse_transform([testY]) # calculate root mean squared error trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:, 0])) print('Train Score: %.2f RMSE' % (trainScore)) testScore = math.sqrt(mean_squared_error(testY[0], testPredict[:, 0])) print('Test Score: %.2f RMSE' % (testScore)) # shift train predictions for plotting trainPredictPlot = numpy.empty_like(dataset) trainPredictPlot[:, :] = numpy.nan trainPredictPlot[look_back:len(trainPredict)+look_back, :] = trainPredict # shift test predictions for plotting testPredictPlot = numpy.empty_like(dataset) testPredictPlot[:, :] = numpy.nan testPredictPlot[ len(trainPredict)+(look_back*2)+1:len(dataset)-1, :] = testPredict # plot baseline and predictions plt.plot(scaler.inverse_transform(dataset), label='Complete Data') plt.plot(trainPredictPlot, label='Training Data') plt.plot(testPredictPlot, label='Prediction Data') plt.legend(loc='upper left') plt.title('Using {} GPUs'.format(gpus)) plt.show() if __name__ == "__main__": main()
I'm a machine learning hobbyist and discovered the source of the issue after a lot of frustration with my results.
I have a question that I hope either of you can answer.
In the @sparkingarthur code for the single GPU, training is done on parallel_model but predicitions are done using the restored serial_model from disk. Is parallel_model just a reference to the original serial_model? If so, wouldn't it work the same if you just did the predictions on parallel_model instead? If not, why?
@palisadoes (fellow machine learning hobbyist), unfortunately I haven't acquired a multi-gpu setup yet (as I mentioned above I was kind of waiting for this issue to be resolved first but at the moment my single watercooled 1080ti works just fine) so I cannot test code for a >1 GPU setup.
In the example from @sparkingarthur the serial model is not trained, I don't see any .fit or .compile for the serial model. However, I assume @sparkingarthur means to study the differences between making predictions on the serial model and one the parallel model (and in this comparison that's OK), and I think these lines do exactly this:
if gpus == 1:
trainPredict = serial_model.predict(trainX)
testPredict = serial_model.predict(testX)
else:
trainPredict = parallel_model.predict(trainX)
testPredict = parallel_model.predict(testX)
I don't think it is possible to a obtain "correct" prediction using a multi_gpu setup for the same reason as why it is not possible to obtain a "correct" prediction when using a batch size > 1 for a stateful LSTM (time series) model (this regardless of the number of GPUs, of course). However, this doesn't mean that there is a more sophisticated way of splitting the data for a multi_gpu_model. It should be possible to improve the get_slice() function, or what do you think?
@palisadoes in the tensorflow2.0 documentation mentioned that multi_gpu functionality is deprecated. The best practice for using multiple GPUs is to use tf.distribute.Strategy. I changed a little bit your code, wherein my two GPUs, it works.
#!/usr/bin/env python3
"""LSTM for sinusoidal data problem with regression framing.
Based on:
https://machinelearningmastery.com/time-series-prediction-lstm-recurrent-neural-networks-python-keras/
"""
# Standard imports
import argparse
import math
# PIP3 imports
import numpy
import matplotlib.pyplot as plt
import tensorflow as tf
from pandas import DataFrame
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.utils import multi_gpu_model
import tensorflow as tf
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error
tf.debugging.set_log_device_placement(True)
# convert an array of values into a dataset matrix
def create_dataset(dataset, look_back=1):
dataX, dataY = [], []
for i in range(len(dataset)-look_back-1):
a = dataset[i:(i+look_back), 0]
dataX.append(a)
dataY.append(dataset[i + look_back, 0])
return numpy.array(dataX), numpy.array(dataY)
def main():
# fix random seed for reproducibility
numpy.random.seed(7)
# Get CLI arguments
parser = argparse.ArgumentParser()
parser.add_argument(
'--gpus',
help='Number of GPUs to use.',
type=int, default=1)
args = parser.parse_args()
gpus = args.gpus
# load the dataset
dataframe = DataFrame(
[0.00000, 5.99000, 11.92016, 17.73121, 23.36510, 28.76553, 33.87855,
38.65306, 43.04137, 46.99961, 50.48826, 53.47244, 55.92235, 57.81349,
59.12698, 59.84970, 59.97442, 59.49989, 58.43086, 56.77801, 54.55785,
51.79256, 48.50978, 44.74231, 40.52779, 35.90833, 30.93008, 25.64279,
20.09929, 14.35496, 8.46720, 2.49484, -3.50245, -9.46474, -15.33247,
-21.04699, -26.55123, -31.79017, -36.71147, -41.26597, -45.40815,
-49.09663, -52.29455, -54.96996, -57.09612, -58.65181, -59.62146,
-59.99540, -59.76988, -58.94716, -57.53546, -55.54888, -53.00728,
-49.93605, -46.36587, -42.33242, -37.87600, -33.04113, -27.87613,
-22.43260, -16.76493, -10.92975, -4.98536, 1.00883, 6.99295, 12.90720,
18.69248, 24.29100, 29.64680, 34.70639, 39.41920, 43.73814, 47.62007,
51.02620, 53.92249, 56.28000, 58.07518, 59.29009, 59.91260, 59.93648,
59.36149, 58.19339, 56.44383, 54.13031, 51.27593, 47.90923, 44.06383,
39.77815, 35.09503, 30.06125, 24.72711, 19.14590, 13.37339, 7.46727,
1.48653, -4.50907, -10.45961, -16.30564, -21.98875, -27.45215,
-32.64127, -37.50424, -41.99248, -46.06115, -49.66959, -52.78175,
-55.36653, -57.39810, -58.85617, -59.72618, -59.99941, -59.67316,
-58.75066, -57.24115, -55.15971, -52.52713, -49.36972, -45.71902,
-41.61151, -37.08823, -32.19438, -26.97885, -21.49376, -15.79391,
-9.93625, -3.97931, 2.01738, 7.99392, 13.89059, 19.64847, 25.21002,
30.51969, 35.52441, 40.17419, 44.42255, 48.22707, 51.54971, 54.35728,
56.62174, 58.32045, 59.43644, 59.95856, 59.88160, 59.20632, 57.93947,
56.09370, 53.68747, 50.74481, 47.29512, 43.37288, 39.01727, 34.27181,
29.18392, 23.80443, 18.18710, 12.38805, 6.46522, 0.47779, -5.51441,
-11.45151])
dataset = dataframe.values
dataset = dataset.astype('float32')
# normalize the dataset
scaler = MinMaxScaler(feature_range=(0, 1))
dataset = scaler.fit_transform(dataset)
# split into train and test sets
train_size = int(len(dataset) * 0.67)
test_size = len(dataset) - train_size
train, test = dataset[0:train_size, :], dataset[train_size:len(dataset), :]
# reshape into X=t and Y=t+1
look_back = 1
trainX, trainY = create_dataset(train, look_back)
testX, testY = create_dataset(test, look_back)
# reshape input to be [samples, time steps, features]
trainX = numpy.reshape(trainX, (trainX.shape[0], 1, trainX.shape[1]))
testX = numpy.reshape(testX, (testX.shape[0], 1, testX.shape[1]))
# create and fit the LSTM network
model = tf.keras.Sequential()
model.add(tf.keras.layers.LSTM(4, input_shape=(1, look_back)))
model.add(tf.keras.layers.Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
if gpus == 1:
model = tf.keras.Sequential()
model.add(tf.keras.layers.LSTM(4, input_shape=(1, look_back)))
model.add(tf.keras.layers.Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
else:
strategy = tf.distribute.MirroredStrategy(devices=["/gpu:0", "/gpu:1"])
print('Number of devices: {}'.format(strategy.num_replicas_in_sync))
# Define the model
with strategy.scope():
model = tf.keras.Sequential()
model.add(tf.keras.layers.LSTM(4, input_shape=(1, look_back)))
model.add(tf.keras.layers.Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
# inputs = tf.keras.layers.Input(shape=(1, look_back))
# lstm_layer = tf.keras.layers.LSTM(4)(inputs)
# outputs = tf.keras.layers.Dense(1)(lstm_layer)
# model = tf.keras.Model(inputs, outputs)
# model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(trainX, trainY, epochs=100, batch_size=4, verbose=2)
trainPredict = model.predict(trainX)
testPredict = model.predict(testX)
# invert predictions
trainPredict = scaler.inverse_transform(trainPredict)
trainY = scaler.inverse_transform([trainY])
testPredict = scaler.inverse_transform(testPredict)
testY = scaler.inverse_transform([testY])
# calculate root mean squared error
trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:, 0]))
print('Train Score: %.2f RMSE' % (trainScore))
testScore = math.sqrt(mean_squared_error(testY[0], testPredict[:, 0]))
print('Test Score: %.2f RMSE' % (testScore))
# shift train predictions for plotting
trainPredictPlot = numpy.empty_like(dataset)
trainPredictPlot[:, :] = numpy.nan
trainPredictPlot[look_back:len(trainPredict)+look_back, :] = trainPredict
# shift test predictions for plotting
testPredictPlot = numpy.empty_like(dataset)
testPredictPlot[:, :] = numpy.nan
testPredictPlot[
len(trainPredict)+(look_back*2)+1:len(dataset)-1, :] = testPredict
# plot baseline and predictions
plt.plot(scaler.inverse_transform(dataset), label='Complete Data')
plt.plot(trainPredictPlot, label='Training Data')
plt.plot(testPredictPlot, label='Prediction Data')
plt.legend(loc='upper left')
plt.title('Using {} GPUs'.format(gpus))
plt.show()
if __name__ == "__main__":
main()
you should use the same total batch size as below
parallel_model.fit(
trainX, trainY,
epochs=100,
batch_size=int(dataset.size / 20),
verbose=2)
or increase learning rate when you use larger batch size as suggested in this paper
Accurate, Large Minibatch SGD
adam = Adam(lr=0.001 * gpus, beta_1=0.9, beta_2=0.999, amsgrad=False)
parallel_model.compile( loss='mean_squared_error', optimizer=adam)
parallel_model.fit(
trainX, trainY,
epochs=100,
batch_size=int(dataset.size * gpus / 20),
verbose=2)
when use 1 GPU and batch_size=int(dataset.size * 4 / 20) it should give the bad result again
I think there are no problems in multi_gpu_model(...)
Thanks @gibrano, I finally got back to looking at this. I made some modifications to your code example. The charts look good for up to 3 of 4 GPUs. (I have a 4 GPU system, so this could be an n-1 issue). Read more below.
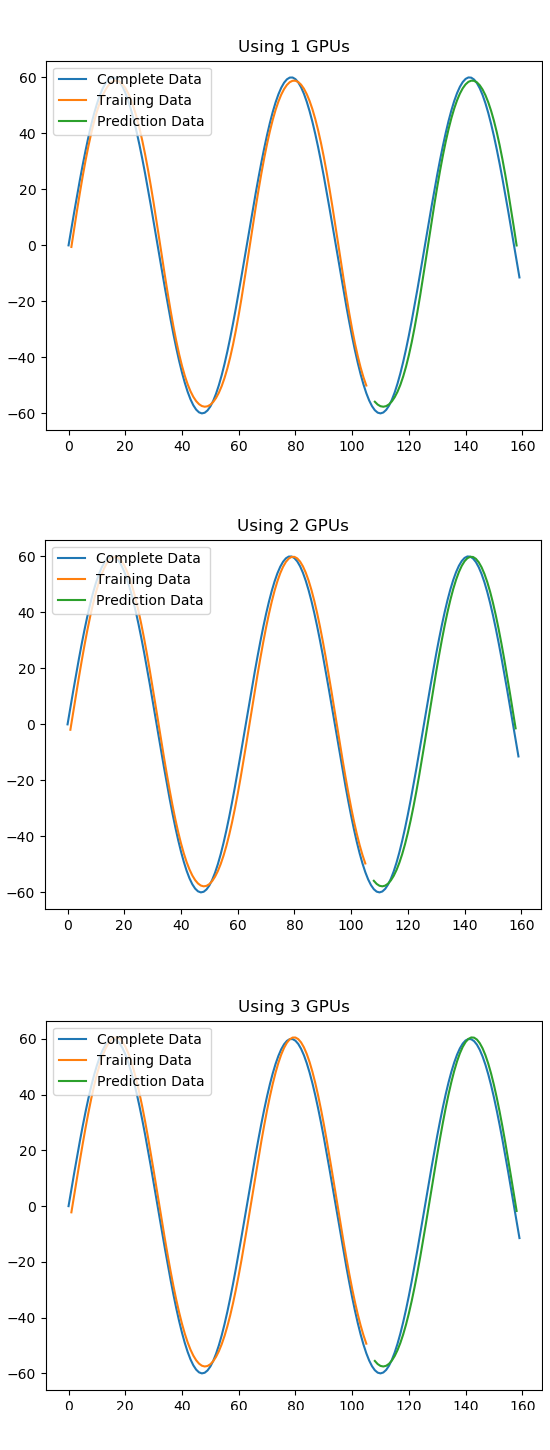
However it fails when running on 4 GPUs (the max on my system). I used the batch size suggestion from @dolaamon2, but it fails for a variety of sizes.
The failure appears to be on the very first epoch run.
gradients/concat_grad/Rank: (Const): /job:localhost/replica:0/task:0/device:GPU:3
gradients/concat_grad/Shape: (Const): /job:localhost/replica:0/task:0/device:GPU:3
gradients/concat_grad/Shape_1: (Const): /job:localhost/replica:0/task:0/device:GPU:3
1/4 [======>.......................] - ETA: 0s - loss: 0.5463Traceback (most recent call last):
File "timeseries/obya/bin/multi-gpu-tester.py", line 219, in <module>
main()
File "timeseries/obya/bin/multi-gpu-tester.py", line 176, in main
model.fit(
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/keras/engine/training.py", line 66, in _method_wrapper
return method(self, *args, **kwargs)
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/keras/engine/training.py", line 848, in fit
tmp_logs = train_function(iterator)
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/def_function.py", line 580, in __call__
result = self._call(*args, **kwds)
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/def_function.py", line 611, in _call
return self._stateless_fn(*args, **kwds) # pylint: disable=not-callable
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/function.py", line 2420, in __call__
return graph_function._filtered_call(args, kwargs) # pylint: disable=protected-access
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/function.py", line 1661, in _filtered_call
return self._call_flat(
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/function.py", line 1745, in _call_flat
return self._build_call_outputs(self._inference_function.call(
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/function.py", line 593, in call
outputs = execute.execute(
File "/usr/local/lib/python3.8/dist-packages/tensorflow/python/eager/execute.py", line 59, in quick_execute
tensors = pywrap_tfe.TFE_Py_Execute(ctx._handle, device_name, op_name,
tensorflow.python.framework.errors_impl.UnknownError: [_Derived_] CUDNN_STATUS_BAD_PARAM
in tensorflow/stream_executor/cuda/cuda_dnn.cc(1459): 'cudnnSetTensorNdDescriptor( tensor_desc.get(), data_type, sizeof(dims) / sizeof(dims[0]), dims, strides)'
[[{{node CudnnRNN}}]]
[[replica_3/sequential/lstm/StatefulPartitionedCall]]
[[div_no_nan/ReadVariableOp_3/_128]] [Op:__inference_train_function_11625]
Function call stack:
train_function -> train_function -> train_function
I'm running:
- Ubuntu 20.04
- Nvidia Driver Version: 440.64
- CUDA Version: 10.2
- tensorflow-gpu Version: 2.2.0
#!/usr/bin/env python3
"""LSTM for sinusoidal data problem with regression framing.
Based on:
https://machinelearningmastery.com/time-series-prediction-lstm-recurrent-neural-networks-python-keras/
"""
# Standard imports
import argparse
import math
from collections import namedtuple
import os
# PIP3 imports
import numpy
import matplotlib.pyplot as plt
import tensorflow as tf
from pandas import DataFrame
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error
tf.debugging.set_log_device_placement(True)
def _device_name(item):
"""Create a device name for Tensorflow.
Args:
item: tensorflow.python.eager.context.PhysicalDevice
Returns:
result: Name of device for tensorflow
"""
# Process
components = item.split(':')
result = '/{}'.format(':'.join(components[1:]))
return result
def setup():
"""Setup TensorFlow 2 operating parameters.
Args:
None
Returns:
result: Processor namedtuple of GPUs and CPUs in the system
"""
# Initialize key variables
Processors = namedtuple('Processors', 'gpus, cpus')
gpu_names = []
cpu_names = []
# Reduce error logging
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
# Limit Tensorflow v2 Limit GPU Memory usage
# https://www.tensorflow.org/guide/gpu
gpus = tf.config.experimental.list_physical_devices('GPU')
cpus = tf.config.experimental.list_physical_devices('CPU')
if bool(gpus) is True:
try:
# Currently, memory growth needs to be the same across GPUs
for gpu in gpus:
gpu_names.append(_device_name(gpu.name))
# Currently, memory growth needs to be the same across GPUs
for _, cpu in enumerate(cpus):
cpu_names.append(_device_name(cpu.name))
except RuntimeError as e:
# Memory growth must be set before GPUs have been initialized
print(e)
# Return
result = Processors(gpus=gpu_names, cpus=cpu_names)
return result
# convert an array of values into a dataset matrix
def create_dataset(dataset, look_back=1):
dataX, dataY = [], []
for i in range(len(dataset)-look_back-1):
a = dataset[i:(i+look_back), 0]
dataX.append(a)
dataY.append(dataset[i + look_back, 0])
return numpy.array(dataX), numpy.array(dataY)
def main():
# Setup memory
processors = setup()
# fix random seed for reproducibility
numpy.random.seed(7)
# Get CLI arguments
parser = argparse.ArgumentParser()
parser.add_argument(
'--gpus',
help='Number of GPUs to use.',
type=int, default=1)
args = parser.parse_args()
_gpus = max(1, abs(args.gpus))
# load the dataset
dataframe = DataFrame(
[0.00000, 5.99000, 11.92016, 17.73121, 23.36510, 28.76553, 33.87855,
38.65306, 43.04137, 46.99961, 50.48826, 53.47244, 55.92235, 57.81349,
59.12698, 59.84970, 59.97442, 59.49989, 58.43086, 56.77801, 54.55785,
51.79256, 48.50978, 44.74231, 40.52779, 35.90833, 30.93008, 25.64279,
20.09929, 14.35496, 8.46720, 2.49484, -3.50245, -9.46474, -15.33247,
-21.04699, -26.55123, -31.79017, -36.71147, -41.26597, -45.40815,
-49.09663, -52.29455, -54.96996, -57.09612, -58.65181, -59.62146,
-59.99540, -59.76988, -58.94716, -57.53546, -55.54888, -53.00728,
-49.93605, -46.36587, -42.33242, -37.87600, -33.04113, -27.87613,
-22.43260, -16.76493, -10.92975, -4.98536, 1.00883, 6.99295, 12.90720,
18.69248, 24.29100, 29.64680, 34.70639, 39.41920, 43.73814, 47.62007,
51.02620, 53.92249, 56.28000, 58.07518, 59.29009, 59.91260, 59.93648,
59.36149, 58.19339, 56.44383, 54.13031, 51.27593, 47.90923, 44.06383,
39.77815, 35.09503, 30.06125, 24.72711, 19.14590, 13.37339, 7.46727,
1.48653, -4.50907, -10.45961, -16.30564, -21.98875, -27.45215,
-32.64127, -37.50424, -41.99248, -46.06115, -49.66959, -52.78175,
-55.36653, -57.39810, -58.85617, -59.72618, -59.99941, -59.67316,
-58.75066, -57.24115, -55.15971, -52.52713, -49.36972, -45.71902,
-41.61151, -37.08823, -32.19438, -26.97885, -21.49376, -15.79391,
-9.93625, -3.97931, 2.01738, 7.99392, 13.89059, 19.64847, 25.21002,
30.51969, 35.52441, 40.17419, 44.42255, 48.22707, 51.54971, 54.35728,
56.62174, 58.32045, 59.43644, 59.95856, 59.88160, 59.20632, 57.93947,
56.09370, 53.68747, 50.74481, 47.29512, 43.37288, 39.01727, 34.27181,
29.18392, 23.80443, 18.18710, 12.38805, 6.46522, 0.47779, -5.51441,
-11.45151])
dataset = dataframe.values
dataset = dataset.astype('float32')
# normalize the dataset
scaler = MinMaxScaler(feature_range=(0, 1))
dataset = scaler.fit_transform(dataset)
# split into train and test sets
train_size = int(len(dataset) * 0.67)
test_size = len(dataset) - train_size
train, test = dataset[0:train_size, :], dataset[train_size:len(dataset), :]
# reshape into X=t and Y=t+1
look_back = 1
trainX, trainY = create_dataset(train, look_back)
testX, testY = create_dataset(test, look_back)
# reshape input to be [samples, time steps, features]
trainX = numpy.reshape(trainX, (trainX.shape[0], 1, trainX.shape[1]))
testX = numpy.reshape(testX, (testX.shape[0], 1, testX.shape[1]))
# Create and fit the LSTM network
if _gpus > len(processors.gpus):
devices = processors.gpus
else:
devices = processors.gpus[:min(_gpus, len(processors.gpus))]
gpus = len(devices)
strategy = tf.distribute.MirroredStrategy(devices=devices)
print('Number of devices: {}'.format(strategy.num_replicas_in_sync))
# Define the model
with strategy.scope():
model = tf.keras.Sequential()
model.add(tf.keras.layers.LSTM(4, input_shape=(1, look_back)))
model.add(tf.keras.layers.Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(
trainX,
trainY,
epochs=500,
batch_size=int(dataset.size * len(devices) / 20),
verbose=1)
trainPredict = model.predict(trainX)
testPredict = model.predict(testX)
# invert predictions
trainPredict = scaler.inverse_transform(trainPredict)
trainY = scaler.inverse_transform([trainY])
testPredict = scaler.inverse_transform(testPredict)
testY = scaler.inverse_transform([testY])
# calculate root mean squared error
trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:, 0]))
print('Train Score: %.2f RMSE' % (trainScore))
testScore = math.sqrt(mean_squared_error(testY[0], testPredict[:, 0]))
print('Test Score: %.2f RMSE' % (testScore))
# shift train predictions for plotting
trainPredictPlot = numpy.empty_like(dataset)
trainPredictPlot[:, :] = numpy.nan
trainPredictPlot[look_back:len(trainPredict)+look_back, :] = trainPredict
# shift test predictions for plotting
testPredictPlot = numpy.empty_like(dataset)
testPredictPlot[:, :] = numpy.nan
testPredictPlot[
len(trainPredict)+(look_back*2)+1:len(dataset)-1, :] = testPredict
# plot baseline and predictions
plt.plot(scaler.inverse_transform(dataset), label='Complete Data')
plt.plot(trainPredictPlot, label='Training Data')
plt.plot(testPredictPlot, label='Prediction Data')
plt.legend(loc='upper left')
plt.title('Using {} GPUs'.format(gpus))
plt.show()
if __name__ == "__main__":
main()
Comparing the logs from 3 vs 4 GPUs, the crash occurs crash right after all the gradients are calculated on all GPUs.
I know Cuda 10-2 isn't recommended, so I'll downgrade and see if the issue persists and will update this thread. If it does, then I'll open another issue, as TensorFlow 2.2 seems to be working otherwise.
Most helpful comment
Thanks @gibrano, I finally got back to looking at this. I made some modifications to your code example. The charts look good for up to 3 of 4 GPUs. (I have a 4 GPU system, so this could be an n-1 issue). Read more below.
However it fails when running on 4 GPUs (the max on my system). I used the batch size suggestion from @dolaamon2, but it fails for a variety of sizes.
The failure appears to be on the very first epoch run.
I'm running:
Comparing the logs from 3 vs 4 GPUs, the crash occurs crash right after all the gradients are calculated on all GPUs.
I know Cuda 10-2 isn't recommended, so I'll downgrade and see if the issue persists and will update this thread. If it does, then I'll open another issue, as TensorFlow 2.2 seems to be working otherwise.