Keras: Passing additional arguments to objective function
Is it possible to pass additional arguments (**kvargs) to a custom objective function?
Specifically I'd like the mse to also maximize the difference between two different target outputs, i.e. something like:
def interference_mean_squared_error(y_true, y_pred, other_y_pred):
return K.mean(K.square(y_pred - y_true) - K.square(y_true - other_y_pred) , axis=-1)
All 29 comments
Alternatively, can we combine two Graph outputs and have them share objective function? Maybe reshaping would work, but it seems cumbersome.
@carlthome I'm also interested in this. In my case, there's an additional cost term depending on the hidden activations at different time steps.
I've done something similar in #1061 using partial function evaluation. The main idea is return another function within your loss function that conforms to the API. It's not pretty but it works.
Thanks @rpinsler! :smile:
If I get your solution correctly it seems you concatenate outputs and slice them inside your loss function? The part I'm most unsure about is the theano.scan inside gmm_loss though. Why is the scan needed? I mean, going by the native mean-square error for example, it doesn't do any looping. Is it because of the slicing inside the nested loss function?
Yes, exactly. Basically my output is the concatenation of M means (D-dimensional), M variances and M weights for the Gaussian components. The scan loop goes from 1:M, indexing the corresponding elements. Probably a better solution would be to fork the outputs (into 3 forks: means, variances, weights) and then apply the different activation functions to all elements of each fork. This should be much easier with the new functional API now as it was before.
Obviously, this is irrelevant to your original question. You only need something like this (untested):
def interference_mean_squared_error(other_y_pred):
def imse(y_true, y_pred):
return K.mean(K.square(y_pred - y_true) - K.square(y_true - other_y_pred) , axis=-1)
return imse
I just realize other_y_pred is some dynamic output as well, so the above solution isn't enough indeed. Should be easier with the new functional API though.
@rpinsler, actually doing a closure in the manner you suggested seems to work (thanks!). I'm running experiments now and will know tomorrow if it converges to something sensible or not. I assume everything's pass-by-reference and that the output tensors are after the activation function. Bit tricky to know for sure though. I'm not particularly comfortable with the backend bridge.
def penalized_loss(noise):
def loss(y_true, y_pred):
return K.mean(K.square(y_pred - y_true) - K.square(y_true - noise), axis=-1)
return loss
input1 = Input(batch_shape=(batch_size, timesteps, features))
lstm = LSTM(features, stateful=True, return_sequences=True)(input1)
output1 = TimeDistributed(Dense(features, activation='sigmoid'))(lstm)
output2 = TimeDistributed(Dense(features, activation='sigmoid'))(lstm)
model = Model(input=[input1], output=[output1, output2])
model.compile(loss=[penalized_loss(noise=output2), penalized_loss(noise=output1)], optimizer='rmsprop')
I assume everything's pass-by-reference and that the output tensors are after the activation function
I am pretty sure this is true. When you create use Input, a symbolic variable is created. Then, the tensor operations are applied to this symbolic variable. And actually, even when you pass in the loss, a symbolic variable is created for y_true.
The wrapping of the function solution is really neat. It solves a couple of issues I was struggling with concerning baking in some online reinforcement learning values into the loss.
by the way, the losses also have some fancy calculations when they get wrapped for weightedness. I see that you are returning the mean loss. I don't know if that'll mean anything for you in this instance, but I thought you should have it pointed out because the code generally assumes that the loss isn't mean'ed yet when it runs your function:
https://github.com/fchollet/keras/blob/master/keras/engine/training.py#L309
Thanks @braingineer! I really appreciate the extra pair of eyes. :smiley:
Yes, I am returning a mean but so do most of the predefined loss functions, thus that shouldn't cause any weirdness, right?
In fact, everything seems to work out fine during training. The loss function is now mostly negative of course, but that's what I want. The distance between the input and target should be as close to zero as possible as per your standard MSE, but the distance between the target and the _other_ output should be as large as possible because I want to minimize signal interference, or crosstalk. (e.g. source separation)
Yes, I am returning a mean but so do most of the predefined loss functions, thus that shouldn't cause any weirdness, right?
I didn't think it would be a problem, but I thought I'd point it out, just in case.
but the distance between the target and the other output should be as large as possible because I want to minimize signal interference, or crosstalk
You may also be interested in cross covariance loss; it can be used to disentangle latent factors.
Arxiv paper. Chainer implementation.
Nice! That looks really promising.
Thanks! This really help me a lot!
@ylqfp neat :smile:
When we have multiple objectives loss. Does keras sum each loss function to compute the resulted loss ?
@msobroza , from a quick look at the compile function from engine/training.py it certainly seems to be the case that multiple losses are added together.
@carlthome
As @msobroza shows, keras sum each loss function to compute the resulted loss. So It looks like your loss will always be equal to 0, as penalized_loss(noise=output2)(output1) is the opposite of penalized_loss(noise=output1)(output2) . Have you noticed a problem of this kind?
This closure technique seems like it is going to be a mess when people try to save and load models. Is there really no way to explicitly encode a loss that depends upon multiple parts of the model (besides concatenating them together, passing them in, and then splitting them apart)?
@jacksonloper You are right. I implemented the closure technique and had errors while loading the model -- in the part where the custom function has to be mentioned in the custom_objective parameter in load_model().
I have implemented the following code for my model:
def train(args):
#Create vision model and add adapt layer
weight_path = args.weight_path
model = load_model(weight_path)
inp = model.input
out = model.layers[-2].output
adapt_layer = Dense(64, activation='sigmoid', name='adapt_layer')(out)
vision_model = Model(inputs=inp, outputs=adapt_layer)
vision_model.summary()
#I want the FC layers of the vision model to be trainable.
for layer in vision_model.layers[:19]:
layer.trainable = False
for layer in vision_model.layers[19:]:
layer.trainable = True
#For labeled data
labeled_input = Input(shape=(1024, 1024, 3), name='labeled_input')
#For unlabeled data
unlabeled_input= Input(shape=(1024, 1024, 3), name='unlabeled_input')
#getting outputs of the adapt layer based on the input data
labeled_adapt_features = vision_model(labeled_input)
unlabeled_adapt_features = vision_model(unlabeled_input)
#predictions will be made only for labeled dataset. hence only labeled_features is considered.
predictions = Dense(19, activation='softmax', name='predictions', activity_regularizer=regularizers.l2(0.01))(labeled_adapt_features)
classification_model = Model(inputs=[labeled_input, unlabeled_input], outputs=[predictions])
def custom_loss(y_true, y_pred):
categorical_crossentropy_loss = K.categorical_crossentropy(y_true, y_pred)
weight = 0.25
sigmas = [1e-6, 1e-5, 1e-4, 1e-3, 1e-2, 1e-1, 1, 5, 10, 15, 20, 25, 30, 35, 100, 1e3, 1e4, 1e5, 1e6]
gaussian_kernel = partial(utils.gaussian_kernel_matrix, sigmas=tf.constant(sigmas))
cost = tf.reduce_mean(gaussian_kernel(labeled_adapt_features, labeled_adapt_features))
cost += tf.reduce_mean(gaussian_kernel(unlabeled_adapt_features, unlabeled_adapt_features))
cost -= 2 * tf.reduce_mean(gaussian_kernel(labeled_adapt_features, unlabeled_adapt_features))
cost = tf.where(cost > 0, cost, 0)
domain_mmd_loss = tf.maximum(1e-4, cost) * weight
return categorical_crossentropy_loss + domain_mmd_loss
#Metric function : to check what is the domain loss.
#It's the same domain loss code implemented in the above function.
def domain_loss(y_true, y_pred):
weight = 0.25
sigmas = [1e-6, 1e-5, 1e-4, 1e-3, 1e-2, 1e-1, 1, 5, 10, 15, 20, 25, 30, 35, 100, 1e3, 1e4, 1e5, 1e6]
gaussian_kernel = partial(utils.gaussian_kernel_matrix, sigmas=tf.constant(sigmas))
cost = tf.reduce_mean(gaussian_kernel(labeled_adapt_features, labeled_adapt_features))
cost += tf.reduce_mean(gaussian_kernel(unlabeled_adapt_features, unlabeled_adapt_features))
cost -= 2 * tf.reduce_mean(gaussian_kernel(labeled_adapt_features, unlabeled_adapt_features))
cost = tf.where(cost > 0, cost, 0)
domain_mmd_loss = tf.maximum(1e-4, cost) * weight
return domain_mmd_loss
#Compiling the model
classification_model.compile(optimizer=SGD(lr=1e-2, decay=1e-6, momentum=0.9, nesterov= True), loss={'predictions':custom_loss}, metrics=['accuracy', domain_loss, 'categorical_crossentropy'])
'vision_model' is a shared model (implemented based on the shared vision model code on https://keras.io/getting-started/functional-api-guide/ ). I feed input values to 'labeled_input' and 'unlabeled_input' through fit_generator and compute the respective features. The goal is to classify the 'labeled_input' data using 'labeled_adapt_features' and also to use 'unlabeled_adapt_features' to compute the domain loss between the labeled data and the unlabeled data.
The above implementation of custom_loss directly uses 'labeled_adapt_features' and 'unlabeled_adapt_features' to compute the domain loss, and the code doesn't throw any error while training. Snippet while training happens:

As you can see from the snippet, the domain loss is being calculated and the overall 'loss' is the sum of 'domain_loss' and 'categorical_crossentropy' loss values - as implemented in 'custom_loss' function.
Although when I do classification_model.summary(), I don't see 'unlabeled_data' in it which makes me think if unlabeled_adapt_features is computing anything at all.
Model summary:
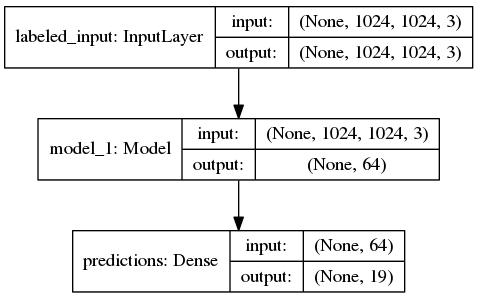
I was expecting to have a model with 2 inputs. I guess a reason 'labeled_input' shows up in the diagram is that I connect 'labeled_adapt_features' to 'predictions' which is an output for the model. Whereas, I compute 'unlabeled_adapt_features' and then do nothing with it in the model due to which I guess it's not showing up. Can anyone tell me if this a correct way to use 'labeled' and 'unlabeled' features in the custom_loss function? Note: Labeled and unlabeled features aren't exactly intermediate layer outputs -- they are the outputs of the vision_model for different set of inputs.
Thanks in advance!
@carlthome , in the back propagation of the loss, the variables of additional arguments(e.g. the output of other layer) will be updated too?
Especially, in the following code which is changed from yours, the variables connected to output2(e.g. Dense layer variables) can be updated in the minimization of the loss like variables connected to output1? In other words, it sees the output2 as a constant tensor or a variable tensor?
def penalized_loss(noise):
def loss(y_true, y_pred):
return K.mean(K.square(y_pred - y_true) - K.square(y_true - noise), axis=-1)
return loss
input1 = Input(batch_shape=(batch_size, timesteps, features))
lstm = LSTM(features, stateful=True, return_sequences=True)(input1)
output1 = TimeDistributed(Dense(features, activation='sigmoid'))(lstm)
output2 = TimeDistributed(Dense(features, activation='sigmoid'))(lstm)
model = Model(input=[input1], output=[output1])
model.compile(loss=[penalized_loss(noise=output2)], optimizer='rmsprop')
@carlthome , I want to use the output of model as well as other layer to define the loss, and the example is as follows:
def loss_fn(other_tensor):
def loss(y_true,y_pred):
return K.sum(K.sqrt(y_pred-other_tensor)+y_pred*y_true)
return loss
inputs = Input(shape)
dense1 = Dense(32)(inputs)
dense2 =Dense(32)(dense1)
dense3 = Dense(16)(dense1)
model = Model(inputs,dense3)
model.compile(optimizer='sgd',loss=loss_fn(dense2))
In the back propagation of the loss, the variables connected to dense2 can be also updated like dense3? In other words, it sees dense2 as constant tensor or variable tensor?
So did anyone solve the questions about loss enclosure mentioned above:
- Does it treat the additional input as a variable or a constant when setting up the gradients?
- Saving models with enclosure?
- How would you perform enclosure with ground truth additional input-- i.e. L(y_true_2, y_pred_2, y_true_1) rather than L(y_true_2, y_pred_2, y_pred_1) where enclosure seemed to work
Along with @MikeShlis, @RohitSaha, and @jacksonloper, I too do not know how to successfully load a model with a loss function defined using the closure method described above.
Along with @MikeShlis, @RohitSaha, and @jacksonloper, I too do not know how to successfully load a model with a loss function defined using the closure method described above.
So i would recommend wrapping keras.models.Model to take your outputs as inputs effectively in keras' eyes, and then using add_loss() to add your loss. Depending on how cleanly you hack this, saving and loading the model will follow the API's normal routine. This also completely frees you in the sense of adding any loss regardless of shape, while enclosure only assists with additional input rather than both input/output.
I was trying to write masked MSE loss:
def mae_loss_masked(mask):
def loss_fn(y_true, y_pred):
abs_vec = tf.multiply(tf.abs(y_pred-y_true), mask)
loss = tf.reduce_mean(abs_vec)
return loss
return loss_fn
My model:
def MobileNet_v1():
# MobileNet with dense layer on top
# Keras 2.1.6
mobilenet = MobileNet(input_shape=(config.IMAGE_H, config.IMAGE_W, config.N_CHANNELS),
alpha=1.0,
depth_multiplier=1,
include_top=False,
weights='imagenet'
)
x = Flatten()(mobilenet.output)
x = Dropout(0.5)(x)
x = Dense(config.N_LANDMARKS * 2, activation='linear')(x)
# -------------------------------------------------------
model = Model(inputs=mobilenet.input, outputs=x)
optimizer = Adadelta()
model.compile(optimizer=optimizer, loss=mae_loss_masked)
model.summary()
import sys
sys.exit()
return model
But it give an error:
TypeError: mae_loss_masked() takes 1 positional argument but 2 were given
Also a question how batch generator should look like in this case.
@mrgloom The loss function you provide must take two arguments, you instead passed in a loss function that only takes one argument (mask). You need to pass in your inner function instead, perhaps it will work if you change loss=mae_loss_masked to loss=mae_loss_masked(yourmask).
@jonathanking Looks like yes, but the problem that my mask is not constant and can be different for each sample.
How are your values masked? If ytrue is padded with a constant value, i.e. 0, you can compute the mask within your loss function.
def loss(ytrue, ypred):
mask=tf.cast(tf.not_equal(ytrue, 0), tf.float32)
ypred=ypred*mask
...
you can pass self.other_pred to your loss function, if self.other_pred is changing every steps, you should
make a variable_scope for it and pass the value in the inputs, make multiple inputs
Most helpful comment
@rpinsler, actually doing a closure in the manner you suggested seems to work (thanks!). I'm running experiments now and will know tomorrow if it converges to something sensible or not. I assume everything's pass-by-reference and that the output tensors are after the activation function. Bit tricky to know for sure though. I'm not particularly comfortable with the backend bridge.