Joss-reviews: [REVIEW]: maxsmooth: Derivative Constrained Function Fitting
Submitting author: @htjb (Harry Bevins)
Repository: https://github.com/htjb/maxsmooth
Version: 1.2.0
Editor: @dfm
Reviewer: @smutch, @benjaminpope
Archive: 10.5281/zenodo.4059338
:warning: JOSS reduced service mode :warning:
Due to the challenges of the COVID-19 pandemic, JOSS is currently operating in a "reduced service mode". You can read more about what that means in our blog post.
Status
Status badge code:
HTML: <a href="https://joss.theoj.org/papers/7f53a67e2a3e8f021d4324de96fb59c8"><img src="https://joss.theoj.org/papers/7f53a67e2a3e8f021d4324de96fb59c8/status.svg"></a>
Markdown: [](https://joss.theoj.org/papers/7f53a67e2a3e8f021d4324de96fb59c8)
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@smutch & @benjaminpope, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
- Make sure you're logged in to your GitHub account
- Be sure to accept the invite at this URL: https://github.com/openjournals/joss-reviews/invitations
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @dfm know.
✨ Please start on your review when you are able, and be sure to complete your review in the next six weeks, at the very latest ✨
Review checklist for @smutch
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@htjb) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
- [x] Substantial scholarly effort: Does this submission meet the scope eligibility described in the JOSS guidelines
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
Review checklist for @benjaminpope
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@htjb) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
- [x] Substantial scholarly effort: Does this submission meet the scope eligibility described in the JOSS guidelines
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
All 67 comments
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @smutch, @benjaminpope it looks like you're currently assigned to review this paper :tada:.
:warning: JOSS reduced service mode :warning:
Due to the challenges of the COVID-19 pandemic, JOSS is currently operating in a "reduced service mode". You can read more about what that means in our blog post.
:star: Important :star:
If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿
To fix this do the following two things:
- Set yourself as 'Not watching' https://github.com/openjournals/joss-reviews:

- You may also like to change your default settings for this watching repositories in your GitHub profile here: https://github.com/settings/notifications

For a list of things I can do to help you, just type:
@whedon commands
For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type:
@whedon generate pdf
Reference check summary:
OK DOIs
- 10.1016/j.physrep.2006.08.002 is OK
- 10.1016/j.physrep.2016.06.006 is OK
- 10.1088/0034-4885/75/8/086901 is OK
- 10.1086/146653 is OK
- 10.1086/106661 is OK
- 10.1093/mnras/stx2065 is OK
- 10.1109/ICEAA.2019.8879199 is OK
- 10.1007/s10686-018-9584-3 is OK
- 10.1093/mnras/sty1244 is OK
- 10.1142/S2251171719500041 is OK
- 10.1038/nature25792 is OK
- 10.3847/1538-4357/aa69bd is OK
- 10.3847/1538-4357/ab2879 is OK
- 10.1021/jp970984n is OK
- 10.1093/comjnl/7.4.308 is OK
MISSING DOIs
- None
INVALID DOIs
- None
Dear @htjb,
Thank you for producing a useful and well-documented software package maxsmooth. I could install and run it with no trouble at all.
Overall it is good and I expect it will not be too much work to fix up such small comments as I have. I will put these here rather than in separate Issues but happy to break any that you think are substantive into Issues.
My first high-level comment is that https://maxsmooth.readthedocs.io is clearer than the paper itself and clarifies some points that I think should be explicitly addressed in the paper. Perhaps overall using some of the text and examples from the readthedocs would help illustrate some points.
About the paper:
- It would be good to say in the paper and early on in the readthedocs that it is written for Python, and which versions it supports.
- The intro paragraph of the readthedocs is very good. In particular, it carries a couple of pieces of information that must go in the paper: specifically, what DCFs are and what they are useful for. When I first read the abstract, I thought, neat, maybe this will be great for fitting out background light in images or light curves, depending on what is meant by derivatives that do not cross zero. But the readthedocs says that this only works for functions with no turning points - so why is it stated as m >=2? Why is this a requirement? I'm just not quite sure what is meant here, and it would be helpful to be more explicit. I tried playing around with the code to allow zero crossings in the first derivative but it won't let me, so I assume this is a feature.
- I think the 'example fit' figure on the GitHub repo is good - perhaps something like this could belong in the paper, with the injected signal overplotted on the residuals so we can see how we went. I have reproduced things like this by myself in python and found it very easy to use so this is great.
About the code/repo:
- Probably something like this would be helpful in 2D for fitting image background gradients - maybe? Is there such a functionality?
- I had no trouble reproducing the examples in the directory example_codes. I notice that these are all Python scripts - but many users are likely to prefer to encounter this in Jupyter Notebooks. Perhaps it would be user-friendly to copy and run these in notebooks?
- If we want to supply our own derivatives, will this play nicely with autodiff libraries like Jax or Theano?
- It would be helpful to have community guidelines for third-party contributors.
About the documentation:
- The intro section (which I've said above is great) is perhaps long and it might be more helpful for users to move the more detailed information and example to below "Installation", which many people will be looking for!
- Under
maxsmoothTheory and Algorithm, it would be helpful to define the terms in the equation explicitly in this section. - In the Parameter Plotter example, there is a cool and vivid corner plot in which invalid regions are grayscale and valid regions are fluro and it looks great. Nevertheless it could do with a paragraph explaining why and how these regions are determined to be valid - I get what you mean, but there is a bit of maths latent in this image that could be brought to the fore.
- In terms of how to use this - I don't have a good intuition for what order N I should go to, or how best to choose a basis. I bet it would be helpful to have examples which illustrate how you would go about choosing this.
I hope that helps! Neat piece of code.
All the best,
Ben
I should add, going back over the checklist - I'm happy to check it off, but it wouldn't hurt to add more description and citation to the State of the Field in the paper.
Dear @benjaminpope,
Thank you for the detailed comments and feedback they are very helpful! I am glad you found the software easy to install and use.
Just as an update, I am currently working my way through your comments and making relevant changes to the paper, docs and repo. I am tracking my changes so that I can make you aware of what I have done and I am aiming to get this sorted ASAP. I will let you know when it is ready for another look.
I have written an extension to one of the functions and just wanted to check that if I push this as version 1.2.0 that will not derail the review process? I of course will update the version number in the paper.
Again thank you for the comments,
Harry
I don't think this would be a problem!
On Thu, Sep 3, 2020 at 6:24 PM htjb notifications@github.com wrote:
Thank you for the detailed comments and feedback they are very helpful! I
am glad you found the software easy to install and use.Just as an update, I am currently working my way through your comments and
making relevant changes to the paper, docs and repo. I am tracking my
changes so that I can make you aware of what I have done and I am aiming to
get this sorted ASAP. I will let you know when it is ready for another look.I have written an extension to one of the functions and just wanted to
check that if I push this as version 1.2.0 that will not derail the review
process? I of course will update the version number in the paper.Again thank you for the comments,
Harry
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_openjournals_joss-2Dreviews_issues_2596-23issuecomment-2D686336432&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=5hrrErX2XoRE9NhfaXas8xzaUjR1mzB1uV0_WcQWjl0&s=yDsaOiQmonYw_PgXROpROnEtfPZOWE1GkvjcWtwmuPk&e=,
or unsubscribe
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_notifications_unsubscribe-2Dauth_ABN6YFJEEIP4S2VBS6THELTSD5HC5ANCNFSM4QJPEWUA&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=5hrrErX2XoRE9NhfaXas8xzaUjR1mzB1uV0_WcQWjl0&s=mSVWFs6LyhEGNY_gWBXwd8wHI1Kzp51UiYsO2KFYmxQ&e=
.
--
Dr Benjamin Pope
NASA Sagan Fellow
Center for Cosmology and Particle Physics // Center for Data Science
New York University
benjaminpope.github.io
@htjb: Don't worry about version numbers re: this review at this point, just handle them however you normally would. I'll walk you through synchronizing the versions at the end.
Hi everyone,
I just wanted to quickly give an apology for my radio silence thus far. I've just returned from annual leave and will be working my way through my review hopefully by the end of this week.
@dfm My invitation has expired. Sorry! Can you please send through a new one.
@whedon re-invite @smutch as reviewer
OK, the reviewer has been re-invited.
@smutch please accept the invite by clicking this link: https://github.com/openjournals/joss-reviews/invitations
Hi @htjb,
This is a really nice, and very useful, package.
First, I'll begin by saying that I agree with all of the feedback already provided by @benjaminpope. I also had no issues installing and using the package and have only minor feedback.
The paper
I have little to add here beyond @benjaminpope's comments. The paper is well written, concise and covers all of the bases.
- It would be nice to have a bit more information for the lay person as to exactly what the package is doing by including the figure from the readme, along with a brief discussion. In the bottom panel of the readme figure it might also be worth plotting the true signal for comparison?
- The speedup of the implemented methodology over the basinhoping method is impressive. It might be nice to include the code you used to generate that plot in the repo as well, for the purposes of reproducability.
The code & documentation
- Some community guidelines on contributing to the code, reporting bugs etc. is required.
- It would be good to make clear in the readme what requirements apply to the pip install and what applies to working with the source (i.e. the wording currently makes it sound like the required packages need to be installed manually / separately).
- In the documentation it would be good to link to the example data wherever appropriate, as readers who just pip installed won't have access to it.
- I found it difficult to ascertain how to fully interpret the
param_plotteroutput. In particular, the valid and invalid regions. Perhaps some extra discussion here would be worth while (as also noted by @benjaminpope). - In the
param_plotteroutput, I notice that the RMS is given in Kelvin. I guess that might not always be the case for all users. :)
Again - I think this is a really nice piece of work and can imagine it getting a lot of use in the 21cm community as well as others where this technique would be applicable.
Cheers,
Simon
Hi @smutch ,
Thank you for the useful comments and feedback. I am glad you were also able to install and use the software.
I have just come back to work after a week off and I am going to continue to work through your comments and @benjaminpope 's also in the comming week.
I am hoping to get back to you with updates by Friday.
Thanks again,
Harry
Dear @benjaminpope, @smutch,
Thanks again for the comments and feedback. I have acted on hopefully all of these now and updated the repository and paper.
Below are some responses to your comments and what I have changed. I have tried to make this as readable as possible, please let me know if something is not clear.
The Paper
Ben
- I have added a note to the paper and docs that the software is written in Python and supports version 3 upwards.
- I have updated the paper/documentation to hopefully clarify some of these points. In terms of the case of a DCF with m >=2 this is a special form of DCF which is referred to as a Maximally Smooth Function or MSF. We can also define a Completely smooth function with m >=1 constrained. I have extended the 'simple_program.py' example to illustrate how we can produce different forms of DCF. It would be great to see maxsmooth used in the context of light curves and it is certainly something I have discussed with colleagues.
- I have added an example demonstration of signal recovery to the paper and pointed the reader to the MNRAS pre-print as well which features many more examples.
Simon
- Again I have added an example use case to the paper. I have also added the true 'signal', noise in this case, to the readme example.
- I have included the code used to compare with the Basin-hopping/Nelder-Mead method into the repo with a note at the end of the README and in the intro to the documentation.
The Code/Repo
Ben
- I am not to sure if this could be implemented in 2D fitting image background gradients because I am unfamiliar with the concept. The code was certainly not designed with this type of application in mind but it would be great to hear your thoughts on it.
- I have included in the repo the examples as Jupyter Notebooks and provided a link to Binder so that they can be used online.
- Community guidelines have been added.
Simon
-I have added a note about the packages that will be installed when installing from source and using the pip install. As far as I am aware all of the requirements apart from those pertaining to documentation and testing will be installed alongside maxsmooth.
The Documentation
Ben
- I have reordered the installation and detailed description in both the README and documentation.
- I have defined the terms in the equation in the Theory and Algorithm section and pointed the reader to the MNRAS pre-print again if they would like to learn more about this.
- I have added to the parameter plotter example a description of the mathematics used to determine whether a region is valid or invalid.
- In terms of choosing a basis and order N of the DCF this is really what the best_basis() function is about and I have added a bit more description about this to the 'best_basis_example.py'. I hope this clarifies this point.
Simon
- I have linked to the example data at the beginning of the maxsmooth examples section for users who have pip installed.
- Again I have added a note to the example on the interpretation of the parameter plotter results.
- Thank you for highlighting the incorrect labelling in the parameter plotter function. I had originally used this to produce graphs for the MNRAS paper and then adapted it into a usable function.
Please let me know if I have missed anything or if there are further comments. As an aside I have released version 1.2.0 and updated the pypi package and repo accordingly. The major difference is an extension to the functionality of the basis_test() function to allow the user to explore other types of DCFs not just MSFs for their fitting problem. I have updated the tests accordingly.
Thanks and looking forward to hearing back,
Harry
I'm sorry human, I don't understand that. You can see what commands I support by typing:
@whedon commands
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
Dear @htjb,
Thanks for the update. I think this is great and am happy to sign off subject to a couple of pretty trivial questions.
I am still a bit confused as to why it has to be monotonic functions. You say you can relax the constraint on the first derivative, so it can cross zero, which I think should allow it to not be monotonic? But you say this just allows for points of inflection. This is just something I'm stuck on conceptually, but maybe it could be good to include a couple of lines of text on page 2 of the paper about this.
The notebooks are great. One thing that bothers me about the examples is... why does it subtract some of the signal in Figure 1? If you choose different parameters can you get different behaviour? It just seems odd to illustrate it with an example where maybe it didn't work as well as in other cases? Anyway otherwise this is fine.
I'm happy with everything otherwise. Well done!
All the best,
Ben
Dear @benjaminpope,
Thank you. There are some responses to your questions below.
- It doesn't need to be monotonic. By default the condition is relaxed on the first derivative and only m>=2 are constrained which allows for a turning point in the data but no inflection points. To allow for inflection points we have to relax the condition on the second derivative as well by changing the 'constraints' keyword argument to 3. I have attached an example figure in which the data has a turning point and we are fitting with a DCF that is constrained with m>=2. I can provide the code for this if you would like to have a look? I hope this clears this point up, please let me know if not. Apologies for any confusion!
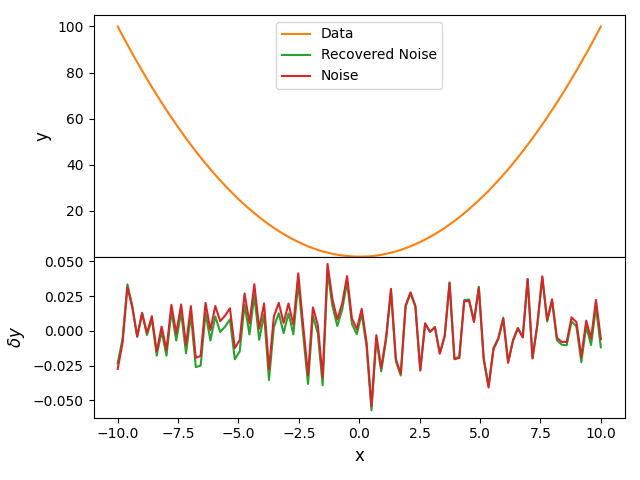
- This is pretty standard behaviour when fitting data with complex non-smooth signals. The recovered signal is actually a 'smooth baseline subtracted' version (I have added this phrase to the paper). Essentially because we are just fitting with a foreground modelled as a DCF that foreground gets distorted by the presence of the signal. The distortion in this case is that the foreground model is 'dragged down' to lower temperatures around 100MHz so that when it is subtracted from the data we actually get a lower amplitude signal. (The distortion is less severe for the README example because the noise is gaussian distributed around 0 so the residuals look better.) If this were a real 21-cm data set then this type of fit would be your initial investigation of the data set. Then because there is evidence of the signal in the residuals you would go onto fit with a signal model and DCF which would result in a better representation of both the foreground and the signal in the data. The final column top row in figure 9 of https://arxiv.org/pdf/2007.14970.pdf shows the residuals in blue when we just subtract a DCF from this exact same data set used for the JOSS example and the orange residuals are when we subtract a DCF and signal model fitted to the data. The recovered signal is then shown in the bottom row same column of this figure in orange next to the actual signal in black. It's a non-trivial point and takes a bit of explaining but I point the reader of the JOSS paper to the MNRAS pre-print for this reason and mention in the caption of the figure that a joint fit results in better representation of the signal.
Hope that helps!
Thanks,
Harry
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
Hi Harry,
This is great, ok. But I guess I got confused by some of the wording and by
trying to run my code (without success) on data with a turn-over - it
failed whenever there was a significant maximum/minimum in the data. I am
sure this is because I ran it incorrectly, but I think it would be nice if
you explicitly stated how to deal with a turning point.
I am sure this is my own misreading of some notes somewhere and apologize
for that.
I also see your second point and am satisfied with the explanation. Happy
to sign off on this review.
All the best,
Ben
On Fri, Sep 18, 2020 at 6:28 PM htjb notifications@github.com wrote:
Thank you. There are some responses to your questions below.
- It doesn't need to be monotonic. By default the condition is relaxed
on the first derivative and only m>=2 are constrained which allows for a
turning point in the data but no inflection points. To allow for inflection
points we have to relax the condition on the second derivative as well by
changing the 'constraints' keyword argument to 3. I have attached an
example figure in which the data has a turning point and we are fitting
with a DCF that is constrained with m>=2. I can provide the code for this
if you would like to have a look? I hope this clears this point up, please
let me know if not. Apologies for any confusion!
[image: turning_point_example]
https://urldefense.proofpoint.com/v2/url?u=https-3A__user-2Dimages.githubusercontent.com_40355093_93572701-2D1e76dd00-2Df98e-2D11ea-2D90fb-2D82a9dac80458.png&d=DwMFaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=932CXALAogGapCzs8SRKY3S9IP_B7DVXBINiAa_VwVA&s=Kel8hQpPEisgU3vfahsxTpcNkxvw4Ywk79rOP-j9fhs&e=- This is pretty standard behaviour when fitting data with complex
non-smooth signals. The recovered signal is actually a 'smooth baseline
subtracted' version (I have added this phrase to the paper). Essentially
because we are just fitting with a foreground modelled as a DCF that
foreground gets distorted by the presence of the signal. The distortion in
this case is that the foreground model is 'dragged down' to lower
temperatures around 100MHz so that when it is subtracted from the data we
actually get a lower amplitude signal. (The distortion is less severe for
the README example because the noise is gaussian distributed around 0 so
the residuals look better.) If this were a real 21-cm data set then this
type of fit would be your initial investigation of the data set. Then
because there is evidence of the signal in the residuals you would go onto
fit with a signal model and DCF which would result in a better
representation of both the foreground and the signal in the data. The final
column top row in figure 9 of https://arxiv.org/pdf/2007.14970.pdf
https://urldefense.proofpoint.com/v2/url?u=https-3A__arxiv.org_pdf_2007.14970.pdf&d=DwMFaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=932CXALAogGapCzs8SRKY3S9IP_B7DVXBINiAa_VwVA&s=57nU7q2d5HgbZ4sx53EGPAcgSty4AcYkDV8ZU6vM8vs&e=
shows the residuals in blue when we just subtract a DCF from this exact
same data set used for the JOSS example and the orange residuals are when
we subtract a DCF and signal model fitted to the data. The recovered signal
is then shown in the bottom row same column of this figure in orange next
to the actual signal in black. It's a non-trivial point and takes a bit of
explaining but I point the reader of the JOSS paper to the MNRAS pre-print
for this reason and mention in the caption of the figure that a joint fit
results in better representation of the signal.Hope that helps!
Thanks,
Harry
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_openjournals_joss-2Dreviews_issues_2596-23issuecomment-2D694733283&d=DwMFaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=932CXALAogGapCzs8SRKY3S9IP_B7DVXBINiAa_VwVA&s=ZL_Id3ImxnRr4xV6bwuaMvIMW9u5Bupl-wZwBJ0NH64&e=,
or unsubscribe
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_notifications_unsubscribe-2Dauth_ABN6YFLOZKZ4JEWKQ7TFUW3SGMK4PANCNFSM4QJPEWUA&d=DwMFaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=932CXALAogGapCzs8SRKY3S9IP_B7DVXBINiAa_VwVA&s=X5A2k1SI0IY1rvMHeQZMA1QBvEr4RS4CUEynIGFSY24&e=
.
--
Dr Benjamin Pope
NASA Sagan Fellow
Center for Cosmology and Particle Physics // Center for Data Science
New York University
benjaminpope.github.io
Hi @benjaminpope,
Thank you. No problem I think it is a bit confusing when you first encounter it and my explanation in places may have been lacking. I will add an example to the docs and notebooks about dealing with turning points and inflection points in data sets which hopefully should make it clearer for first time users.
Thank you for all your comments and feedback!
Best,
Harry
Hi @htjb,
Thanks, great. Subject to that, I sign off - congratulations on a good paper.
All the best,
Ben
Hi @benjaminpope,
I have added that example. Thank you very much again!
Harry
:wave: @smutch: Can you take another look at this when you get a chance to see if your remaining concerns have been addressed or if you have other suggestions? Thanks!
Hi @htjb,
Sorry for the delayed reply (again). I had to take last minute leave last week...
I'm very happy with the changes and improvements. I think it's looking great!
I do have one minor suggestion below but, regardless of this, I am very happy to sign-off on the review:
Since the plotting functions save to a PDF and do not return the underlying matplotlib figure object, the plots do not appear in the example notebooks. You could add something like the following in the notebooks to allow users to see the plots without you having to modify the code:
from IPython.display import IFrame
IFrame("./examples/chi_distribution.pdf", width=800, height=600)
Congrats on a nice package @htjb!
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@whedon check references
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1016/j.physrep.2006.08.002 is OK
- 10.1016/j.physrep.2016.06.006 is OK
- 10.1088/0034-4885/75/8/086901 is OK
- 10.1086/146653 is OK
- 10.1086/106661 is OK
- 10.1093/mnras/stx2065 is OK
- 10.1109/ICEAA.2019.8879199 is OK
- 10.1007/s10686-018-9584-3 is OK
- 10.1093/mnras/sty1244 is OK
- 10.1142/S2251171719500041 is OK
- 10.1038/nature25792 is OK
- 10.3847/1538-4357/aa69bd is OK
- 10.3847/1538-4357/ab2879 is OK
- 10.1021/jp970984n is OK
- 10.1093/comjnl/7.4.308 is OK
MISSING DOIs
- None
INVALID DOIs
- None
@smutch & @benjaminpope: Thanks for your reviews!! I really appreciate your contributions and I believe that your suggestions have helped improve the clarity and impact of the paper and documentation.
@htjb: It looks like we're in good shape here. The only change I would request for the paper is that in the statement of need it says "supports version >=3 [of Python]", but only versions 3.6 and above are tested. I think it would be better to be more specific about the version requirements. After making that change, can you do the following steps and report back here:
- Take a proof read through the manuscript and make sure that you're happy with the proofs - it is harder to make changes later!
- Bump the version number of the software and report that version number back here.
- Archive that release on Zenodo or something similar and post the DOI for that archive to this thread. The metadata (title and author list) for the archive should exactly match this manuscript (you can generally edit the metadata after creating the archive if needed).
Let me know when you've gone through this and we can proceed with acceptance!
@whedon generate pdf
:point_right::page_facing_up: Download article proof :page_facing_up: View article proof on GitHub :page_facing_up: :point_left:
@smutch Thank you for your comments again. I have edited the notebooks so that they include the missing plots.
@dfm I have updated the paper with the appropriate supported python version >=3.6 and proof read through the manuscript. I made one minor change to the wording of one sentence.
The version numbering should be at 1.2.0 on the repository, documentation ect now.
I have also uploaded to Zenodo and the corresponding DOI is 10.5281/zenodo.4059338. Let me know if any of the metadata on this archive needs changing.
Thanks,
Harry
@htjb: Thanks! Can you please update the Zenodo metadata to have your full name as author instead of just your GitHub username?
@whedon set 1.2.0 as version
OK. 1.2.0 is the version.
@whedon set 10.5281/zenodo.4059338 as archive
OK. 10.5281/zenodo.4059338 is the archive.
@dfm Thanks for highlighting this I have changed the author on Zenodo to my full name.
Great! I'll pass this off to the editors in chief for final processing.
@htjb: Congrats and thanks for your submission!
@smutch, @benjaminpope: Thanks again for your reviews!
@whedon accept
Attempting dry run of processing paper acceptance...
:wave: @openjournals/joss-eics, this paper is ready to be accepted and published.
Check final proof :point_right: https://github.com/openjournals/joss-papers/pull/1766
If the paper PDF and Crossref deposit XML look good in https://github.com/openjournals/joss-papers/pull/1766, then you can now move forward with accepting the submission by compiling again with the flag deposit=true e.g.
@whedon accept deposit=true
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1016/j.physrep.2006.08.002 is OK
- 10.1016/j.physrep.2016.06.006 is OK
- 10.1088/0034-4885/75/8/086901 is OK
- 10.1086/146653 is OK
- 10.1086/106661 is OK
- 10.1093/mnras/stx2065 is OK
- 10.1109/ICEAA.2019.8879199 is OK
- 10.1007/s10686-018-9584-3 is OK
- 10.1093/mnras/sty1244 is OK
- 10.1142/S2251171719500041 is OK
- 10.1038/nature25792 is OK
- 10.3847/1538-4357/aa69bd is OK
- 10.3847/1538-4357/ab2879 is OK
- 10.1021/jp970984n is OK
- 10.1093/comjnl/7.4.308 is OK
MISSING DOIs
- None
INVALID DOIs
- None
@dfm, @smutch, @benjaminpope Thank you all for all your help and useful comments!
No worries - congratulations on the acceptance.
On Wed, 30 Sep 2020 at 8:50 pm, htjb notifications@github.com wrote:
>
>
@dfm
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_dfm&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=Ac7io19saHtLg8C6N-_2LPzu12Z7VELr4JPtYTjIF8M&s=fCqHEwbtiptIkSzSt1Vca8DWP0flA0wRJF0DyWVUIv4&e=,
@smutch
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_smutch&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=Ac7io19saHtLg8C6N-_2LPzu12Z7VELr4JPtYTjIF8M&s=klXm2zlKqOkIxoq0xzyFnUhEkyUlZXjvPlEqLX08eqE&e=,
@benjaminpope
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_benjaminpope&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=Ac7io19saHtLg8C6N-_2LPzu12Z7VELr4JPtYTjIF8M&s=0nq0krOTLSVarG-g1y4QrnATqy0XxtXd4FAry6lr9PU&e=
Thank you all for all your help and useful comments!—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_openjournals_joss-2Dreviews_issues_2596-23issuecomment-2D701314269&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=Ac7io19saHtLg8C6N-_2LPzu12Z7VELr4JPtYTjIF8M&s=eS5XRrz9-pbcx7st5OI8Why7UyXlz3A06dKjj5bQEuA&e=,
or unsubscribe
https://urldefense.proofpoint.com/v2/url?u=https-3A__github.com_notifications_unsubscribe-2Dauth_ABN6YFPVMXZVCF47TRU6T2DSIMEN7ANCNFSM4QJPEWUA&d=DwMCaQ&c=slrrB7dE8n7gBJbeO0g-IQ&r=_gjp43gRuTt_LjqJbH0jcZo0ePCz10Y4KegSkx-Ha0A&m=Ac7io19saHtLg8C6N-_2LPzu12Z7VELr4JPtYTjIF8M&s=VGL7gZITehLo-Ghi7ngRVPJdpupurWS6BMs1YNVNuqs&e=
.--
Dr Benjamin Pope
NASA Sagan Fellow
Center for Cosmology and Particle Physics // Center for Data Science
New York University
benjaminpope.github.io
Congrats! 👍
@dfm thanks for editing this work. Perhaps you missed it but the ZENODO archive title does not match the paper, I'll flag this with the author but for future reference try to check this before recommending acceptance.
@htjb I'll process this work for acceptance in JOSS. Below are some last minor points to work on:
[x] Please edit the ZENODO archive metadata so that the title there matches that of the paper
[x] Please write out UK as United Kingdom in your affiliation.
[x] This work is about to be processed for acceptance in JOSS. We recommend that you proofread the paper once more yourself. In particular, ensure the author names and affiliations are correct.
Kevin Moerman
_AEiC JOSS_
@Kevin-Mattheus-Moerman Really? It looked (and looks) like it does to me!
@dfm yes, it is a minor difference but the archive says: htjb/maxsmooth: maxsmooth: Derivative Constrained Function Fitting, while the paper says: maxsmooth: Derivative Constrained Function Fitting, so they need to remove htjb/
@Kevin-Mattheus-Moerman: you're right! I must have interpolated over the repo name every time I looked at it. Sorry!
@Kevin-Mattheus-Moerman Thanks for highlighting the last few points. I have corrected the title on the Zenodo archive, apologies for the error! I have also changed UK to United Kingdom and proof read back through the paper one last time. I have corrected one spelling mistake that I had missed and updated the repo. I am happy for it to go ahead now.
Thanks again,
Harry
@whedon accept
Attempting dry run of processing paper acceptance...
Reference check summary (note 'MISSING' DOIs are suggestions that need verification):
OK DOIs
- 10.1016/j.physrep.2006.08.002 is OK
- 10.1016/j.physrep.2016.06.006 is OK
- 10.1088/0034-4885/75/8/086901 is OK
- 10.1086/146653 is OK
- 10.1086/106661 is OK
- 10.1093/mnras/stx2065 is OK
- 10.1109/ICEAA.2019.8879199 is OK
- 10.1007/s10686-018-9584-3 is OK
- 10.1093/mnras/sty1244 is OK
- 10.1142/S2251171719500041 is OK
- 10.1038/nature25792 is OK
- 10.3847/1538-4357/aa69bd is OK
- 10.3847/1538-4357/ab2879 is OK
- 10.1021/jp970984n is OK
- 10.1093/comjnl/7.4.308 is OK
MISSING DOIs
- None
INVALID DOIs
- None
:wave: @openjournals/joss-eics, this paper is ready to be accepted and published.
Check final proof :point_right: https://github.com/openjournals/joss-papers/pull/1768
If the paper PDF and Crossref deposit XML look good in https://github.com/openjournals/joss-papers/pull/1768, then you can now move forward with accepting the submission by compiling again with the flag deposit=true e.g.
@whedon accept deposit=true
@whedon accept deposit=true
Doing it live! Attempting automated processing of paper acceptance...
🐦🐦🐦 👉 Tweet for this paper 👈 🐦🐦🐦
🚨🚨🚨 THIS IS NOT A DRILL, YOU HAVE JUST ACCEPTED A PAPER INTO JOSS! 🚨🚨🚨
Here's what you must now do:
- Check final PDF and Crossref metadata that was deposited :point_right: https://github.com/openjournals/joss-papers/pull/1769
- Wait a couple of minutes to verify that the paper DOI resolves https://doi.org/10.21105/joss.02596
- If everything looks good, then close this review issue.
Party like you just published a paper! 🎉🌈🦄💃👻🤘
Any issues? Notify your editorial technical team...
:tada::tada::tada: Congratulations on your paper acceptance! :tada::tada::tada:
If you would like to include a link to your paper from your README use the following code snippets:
Markdown:
[](https://doi.org/10.21105/joss.02596)
HTML:
<a style="border-width:0" href="https://doi.org/10.21105/joss.02596">
<img src="https://joss.theoj.org/papers/10.21105/joss.02596/status.svg" alt="DOI badge" >
</a>
reStructuredText:
.. image:: https://joss.theoj.org/papers/10.21105/joss.02596/status.svg
:target: https://doi.org/10.21105/joss.02596
This is how it will look in your documentation:
We need your help!
Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
- Volunteering to review for us sometime in the future. You can add your name to the reviewer list here: https://joss.theoj.org/reviewer-signup.html
- Making a small donation to support our running costs here: https://numfocus.org/donate-to-joss
Congratulations @htjb! Also thank you @smutch, @benjaminpope for reviewing this work and to @dfm for editing it. :tada:
@Kevin-Mattheus-Moerman Thank you very much! And thanks again @dfm, @smutch and @benjaminpope for all your help, the comments were very appreciated!
Most helpful comment
Congratulations @htjb! Also thank you @smutch, @benjaminpope for reviewing this work and to @dfm for editing it. :tada: