Joss-reviews: [REVIEW]: xrnet: Hierarchical Regularized Regression to Incorporate External Data
Submitting author: @gmweaver (Garrett Weaver)
Repository: https://github.com/USCbiostats/xrnet
Version: v0.1.3
Editor: @mgymrek
Reviewer: @juanvillada, @jsgalan
Archive: 10.5281/zenodo.3564788
Status
Status badge code:
HTML: <a href="https://joss.theoj.org/papers/5e290cb57b61f83de4460fd0eca22726"><img src="https://joss.theoj.org/papers/5e290cb57b61f83de4460fd0eca22726/status.svg"></a>
Markdown: [](https://joss.theoj.org/papers/5e290cb57b61f83de4460fd0eca22726)
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@juanvillada & @jsgalan, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
- Make sure you're logged in to your GitHub account
- Be sure to accept the invite at this URL: https://github.com/openjournals/joss-reviews/invitations
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @mgymrek know.
✨ Please try and complete your review in the next two weeks ✨
Review checklist for @juanvillada
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@gmweaver) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
Review checklist for @jsgalan
Conflict of interest
- [x] I confirm that I have read the JOSS conflict of interest (COI) policy and that: I have no COIs with reviewing this work or that any perceived COIs have been waived by JOSS for the purpose of this review.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Contribution and authorship: Has the submitting author (@gmweaver) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the functionality of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Summary: Has a clear description of the high-level functionality and purpose of the software for a diverse, non-specialist audience been provided?
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] State of the field: Do the authors describe how this software compares to other commonly-used packages?
- [x] Quality of writing: Is the paper well written (i.e., it does not require editing for structure, language, or writing quality)?
- [x] References: Is the list of references complete, and is everything cited appropriately that should be cited (e.g., papers, datasets, software)? Do references in the text use the proper citation syntax?
All 49 comments
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @juanvillada, @jsgalan it looks like you're currently assigned to review this paper :tada:.
:star: Important :star:
If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿
To fix this do the following two things:
- Set yourself as 'Not watching' https://github.com/openjournals/joss-reviews:

- You may also like to change your default settings for this watching repositories in your GitHub profile here: https://github.com/settings/notifications

For a list of things I can do to help you, just type:
@whedon commands
For example, to regenerate the paper pdf after making changes in the paper's md or bib files, type:
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
Hi all!
This is a very useful piece of software accompanied by a very well written paper.
Some comments:
⚠️ When I tried to install the Master branch of xrnet on macOS (Mojave 10.14.6) I ran into the error:
ld: warning: directory not found for option '-L/usr/local/gfortran/lib'
✅ No problem with Master branch installation of xrnet in Linux (Ubuntu 19.04)
✅ I am attaching the markdown file from the tests I did on the software. https://www.dropbox.com/s/j0x16cdmdzq7976/xrnet_run.html?dl=0
Thanks to the authors for open sourcing this tool! 👏🏼👏🏼
Hi @juanvillada,
Thanks for the review!
With regard to installation on macOS, I believe this is specific to recent R versions that require a more recent version of GNU Fortran to compile packages from source. See https://cloud.r-project.org/bin/macosx/tools/ for a more detailed description and a binary to install the latest GNU Fortran, which should fix the installation issue. I can also update our documentation to inform users of this detail as well.
Many thanks again for your feedback!
Hi @gmweaver,
Everything works fine after installing the latest GNU Fortran.
✅ No problem with Master branch installation of xrnet on macOS (Mojave 10.14.6).
Best wishes and thanks for this tool!
Thanks @juanvillada for the review!
@jsgalan Please post any comments about your review and complete the checklist above as soon as you are able.
Hi guys, this review escaped from email/alerts/calendars/todo-list. I am sorry.
The article explain the context very well and summarize the need for this tool and how it adds to the existing software packages.
- authors need to explain the acronyms SCAD and MCP or add some references that explains these methods.
Firstly I am using, R version 3.6.0 (2019-04-26) Platform: x86_64-apple-darwin15.6.0 (64-bit) on a MacOS Mojave 10.14.6
I tried the installation via master branch and got 17 warnings plus an installation error, see the resulting error
xrnet.txt
After installing the GNUfortran, as suggested by @gmweaver, I still got 17 warnings but a clean install, see the warnings here
xrnet-gfortran.txt
It finally installed and I ran the demo script.
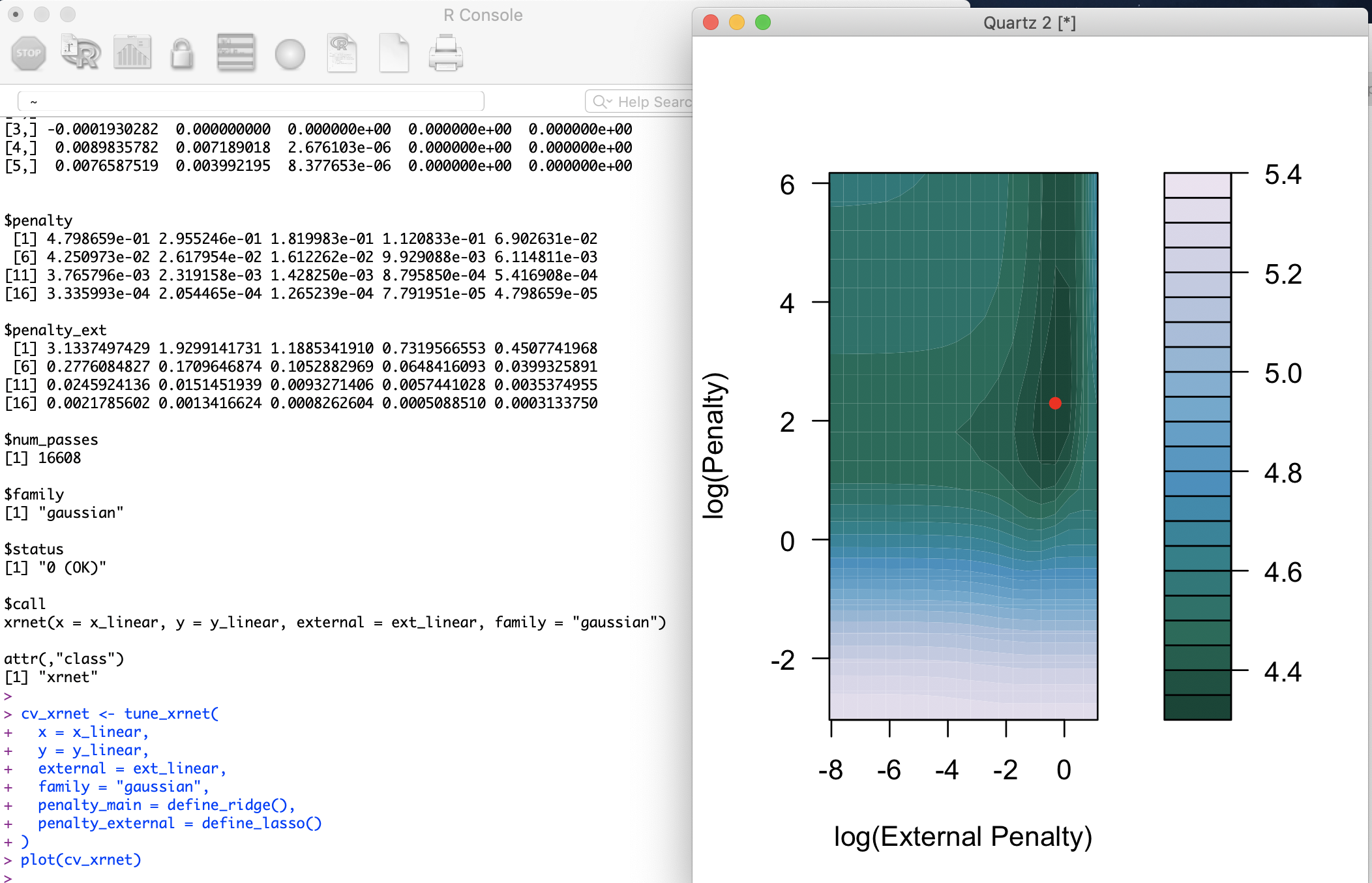
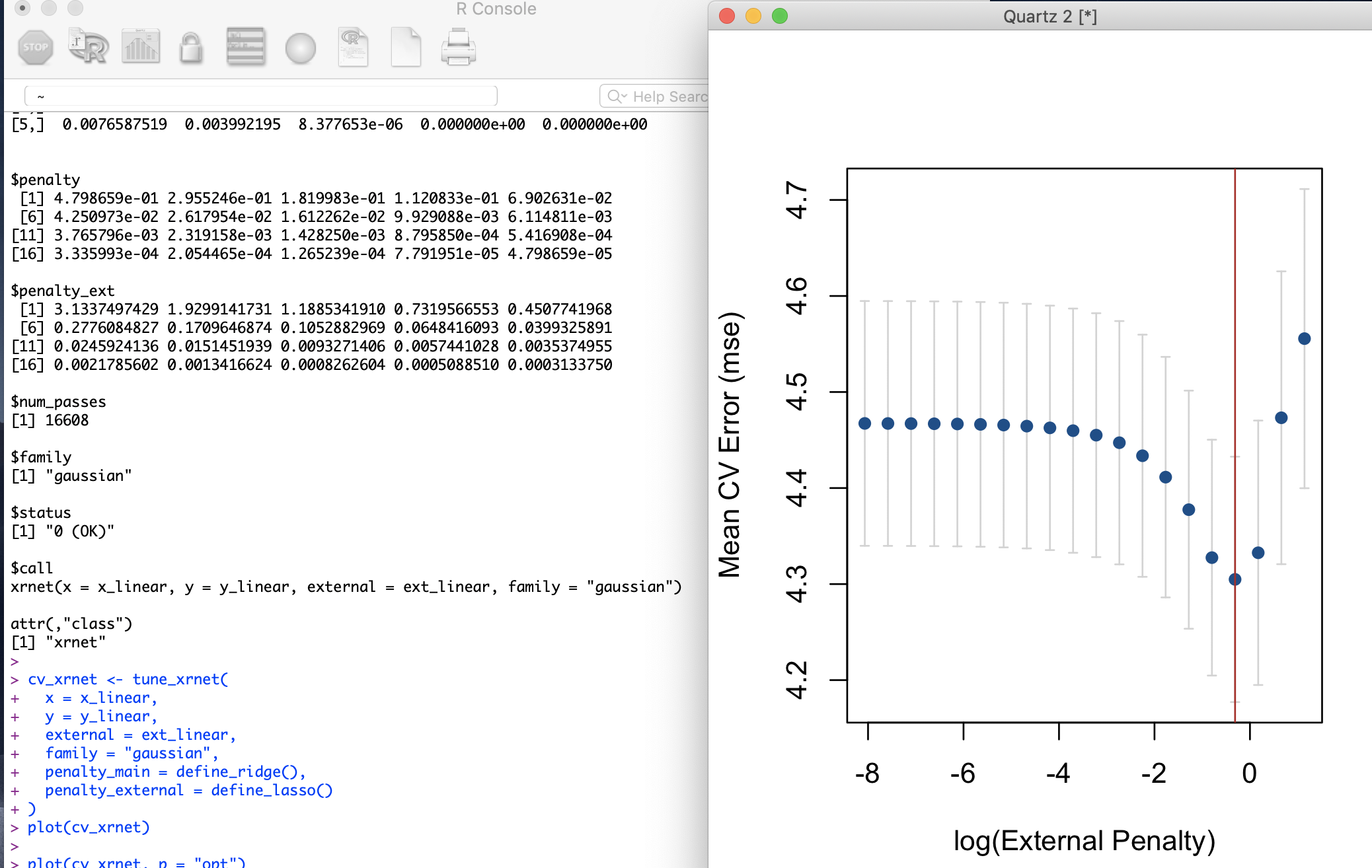
I have a few suggestions for the authors:
- add the GNUfortran installation part of the Setup in the front page of the package repository
- add a second and more complex example, for instance one reading a csv file.
Best,
Thanks @jsgalan and @juanvillada for the reviews.
@gmweaver, can you post your response to these comments regarding the installation process here?
@jsgalan thanks for the review and suggestions!
I have made the following changes based on your feedback:
- Added citations for the SCAD and MCP penalties to the paper
- Added more detail to the installation instructions regarding GNUFortran for Mac
- Added a second example that demonstrates how to use a big.matrix in the package by loading data from a text file. Let me know if this is enough of an addition.
The warnings when installing are specific to RcppEigen and although annoying, should be harmless. See https://stackoverflow.com/questions/49525561/rcppeigen-package-pragma-clang-diagnostic-pop-warnings for notes from one of the maintainers of that package (Dirk Eddelbeuttel)
Hi @gmweaver,
- Please recompile the article via Whedon (whedon generate pdf) to see the changes.
- I checked the updated installation information, its way better.
- On the second example using big.matrix, is "x_linear.txt" a real file? could you provide an example of such file, even if its just 100,000 x 100,000 of random numbers?
Best,
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@jsgalan "x_linear.txt" is a real file stored under inst/extdata in the repository (I followed the best practices suggested in the R Packages book by Hadley Wickham for including raw data files). I have updated the readme to explain where this file is located and and how to access within R after installing the package. Let me know if this will work or if you had something else in mind for a file example.
hey @gmweaver that sounds great, thanks for the explanation. Also, the article checks great.
I am happy with the review.
@jsgalan awesome, thanks for all the feedback!
@mgymrek I believe I have addressed both reviewers comments, let me know if there is anything else I can provide for the review
Great. Thanks @jsgalan and @juanvillada for the reviews and @gmweaver for addressing all comments.
@gmweaver can you fix this minor typo in the paper? Everything else looks good:
"meta-features, also know as" -> "meta-features, also known as"
Then, please make a tagged release and archive (e.g. Zenodo) and report the version number and archive DOI here. Please ensure the title and author list match that in the paper.
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@mgymrek thanks for catching that typo! Fixed and generated the PDF again. I have also released a tagged version and archived to Zenodo.
The version is v0.1.1 and the Zenodo DOI is 10.5281/zenodo.3538245
@mgymrek small update to ensure DESCRIPTION file version matches tagged release version.
New version is v0.1.2 and Zenodo DOI is 10.5281/zenodo.3538278
@whedon set v0.1.2 as version
OK. v0.1.2 is the version.
@whedon set 10.5281/zenodo.3538278 as archive
OK. 10.5281/zenodo.3538278 is the archive.
Thanks @gmweaver for the changes
@openjournals/joss-eics we are ready to accept this article!
Hi - sorry, this doesn't seem to have been acted on by the AEiC on duty at the time, so I will do it now.
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
👋 @gmweaver - There are a few small issues with the paper, as indicated in https://github.com/USCbiostats/xrnet/pull/32. Please merge this, or let me know which changes you disagree with.
@danielskatz reviewed the changes in your pull request and everything looks good! Merged pull request and created a new release with changes.
New version is v0.1.3 and Zenodo DOI is 10.5281/zenodo.3564788
@whedon set v0.1.3 as version
OK. v0.1.3 is the version.
@whedon set 10.5281/zenodo.3564788 as archive
OK. 10.5281/zenodo.3564788 is the archive.
@whedon accept
Attempting dry run of processing paper acceptance...
```Reference check summary:
OK DOIs
- 10.18637/jss.v033.i01 is OK
- doi:10.1214/10-AOAS388 is OK
- 10.18637/jss.v055.i14 is OK
- 10.1198/016214501753382273 is OK
- 10.1214/09-AOS729 is OK
- 10.18637/jss.v052.i05 is OK
- 10.18637/jss.v040.i08 is OK
MISSING DOIs
- None
INVALID DOIs
- None
```
Check final proof :point_right: https://github.com/openjournals/joss-papers/pull/1160
If the paper PDF and Crossref deposit XML look good in https://github.com/openjournals/joss-papers/pull/1160, then you can now move forward with accepting the submission by compiling again with the flag deposit=true e.g.
@whedon accept deposit=true
@whedon accept deposit=true
Doing it live! Attempting automated processing of paper acceptance...
🐦🐦🐦 👉 Tweet for this paper 👈 🐦🐦🐦
🚨🚨🚨 THIS IS NOT A DRILL, YOU HAVE JUST ACCEPTED A PAPER INTO JOSS! 🚨🚨🚨
Here's what you must now do:
- Check final PDF and Crossref metadata that was deposited :point_right: https://github.com/openjournals/joss-papers/pull/1161
- Wait a couple of minutes to verify that the paper DOI resolves https://doi.org/10.21105/joss.01761
- If everything looks good, then close this review issue.
Party like you just published a paper! 🎉🌈🦄💃👻🤘
Any issues? notify your editorial technical team...
Thanks to @juanvillada, @jsgalan for reviewing, and @mgymrek for editing!
And congratulations to @gmweaver and co-author!
:tada::tada::tada: Congratulations on your paper acceptance! :tada::tada::tada:
If you would like to include a link to your paper from your README use the following code snippets:
Markdown:
[](https://doi.org/10.21105/joss.01761)
HTML:
<a style="border-width:0" href="https://doi.org/10.21105/joss.01761">
<img src="https://joss.theoj.org/papers/10.21105/joss.01761/status.svg" alt="DOI badge" >
</a>
reStructuredText:
.. image:: https://joss.theoj.org/papers/10.21105/joss.01761/status.svg
:target: https://doi.org/10.21105/joss.01761
This is how it will look in your documentation:
We need your help!
Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
- Volunteering to review for us sometime in the future. You can add your name to the reviewer list here: https://joss.theoj.org/reviewer-signup.html
- Making a small donation to support our running costs here: https://numfocus.org/donate-to-joss
and sorry for the delay in processing this...
Most helpful comment
Hi @juanvillada,
Thanks for the review!
With regard to installation on macOS, I believe this is specific to recent R versions that require a more recent version of GNU Fortran to compile packages from source. See https://cloud.r-project.org/bin/macosx/tools/ for a more detailed description and a binary to install the latest GNU Fortran, which should fix the installation issue. I can also update our documentation to inform users of this detail as well.
Many thanks again for your feedback!