Joss-reviews: [REVIEW]: xml2jupyter: Mapping parameters between XML and Jupyter widgets
Submitting author: @rheiland (Randy Heiland)
Repository: https://github.com/rheiland/xml2jupyter
Version: v1.0.0
Editor: @lheagy
Reviewers: @choldgraf, @lheagy
Archive: 10.5281/zenodo.3261541
Status
Status badge code:
HTML: <a href="http://joss.theoj.org/papers/447aaae5bcf46877d9722b7ffa6291e2"><img src="http://joss.theoj.org/papers/447aaae5bcf46877d9722b7ffa6291e2/status.svg"></a>
Markdown: [](http://joss.theoj.org/papers/447aaae5bcf46877d9722b7ffa6291e2)
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) by leaving comments in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@choldgraf, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
- Make sure you're logged in to your GitHub account
- Be sure to accept the invite at this URL: https://github.com/openjournals/joss-reviews/invitations
The reviewer guidelines are available here: https://joss.readthedocs.io/en/latest/reviewer_guidelines.html. Any questions/concerns please let @lheagy know.
✨ Please try and complete your review in the next two weeks ✨
Review checklist for @choldgraf
Conflict of interest
- [x] As the reviewer I confirm that I have read the JOSS conflict of interest policy and that there are no conflicts of interest for me to review this work.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Version: Does the release version given match the GitHub release (v1.0.0)?
- [x] Authorship: Has the submitting author (@rheiland) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the function of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Authors: Does the
paper.mdfile include a list of authors with their affiliations? - [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] References: Do all archival references that should have a DOI list one (e.g., papers, datasets, software)?
Review checklist for @lheagy
Conflict of interest
- [x] As the reviewer I confirm that I have read the JOSS conflict of interest policy and that there are no conflicts of interest for me to review this work.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Version: Does the release version given match the GitHub release (v1.0.0)?
- [x] Authorship: Has the submitting author (@rheiland) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the function of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Authors: Does the
paper.mdfile include a list of authors with their affiliations? - [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] References: Do all archival references that should have a DOI list one (e.g., papers, datasets, software)?
All 102 comments
Hello human, I'm @whedon, a robot that can help you with some common editorial tasks. @choldgraf it looks like you're currently assigned as the reviewer for this paper :tada:.
:star: Important :star:
If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿
To fix this do the following two things:
- Set yourself as 'Not watching' https://github.com/openjournals/joss-reviews:

- You may also like to change your default settings for this watching repositories in your GitHub profile here: https://github.com/settings/notifications

For a list of things I can do to help you, just type:
@whedon commands
Attempting PDF compilation. Reticulating splines etc...
Article proof is acceptable to me, just slightly strange with Fig 1 appearing in the middle of an XML block and a couple of line breaks in quoted_strings.
👋 Hi @choldgraf, thanks for being willing to review 🎉: If you haven't yet accepted the JOSS invite, please do so here: https://github.com/openjournals/joss-reviews/invitations. This will let you check off the boxes in the issue thread above. You will also probably want to set yourself as "not watching" this repo so that you don't get flooded with all of the JOSS-review traffic.
As for the review, there is a checklist above to help guide the process. Please feel free to ping me with any questions!
Hey all - I took a pass through the repository and have a few comments and thoughts! I've opened issues for each, will take another pass as things get addressed etc!
I'll update this comment periodically as things change.
Issues added to repository
Blockers
- [x] Installation instructions: https://github.com/rheiland/xml2jupyter/issues/1
- [x] Improved context / purpose of project: https://github.com/rheiland/xml2jupyter/issues/2
- [x] More simplified example/usage demo: https://github.com/rheiland/xml2jupyter/issues/3
- [x] Uncomment test code: https://github.com/rheiland/xml2jupyter/issues/4
Non-blockers
- [x] Improvements to contributing guidelines: https://github.com/rheiland/xml2jupyter/issues/5
Many thanks for your review @choldgraf - these are constructive points.
@rheiland - please take a look through the comments above and keep us posted on your progress
:wave: Hi @rheiland, just checking in. Please ping here when you have had a chance to address @choldgraf's comments above
Hi @lheagy and apologies for taking so long to reply! Thanks for keeping things moving along and thanks to @choldgraf for his initial review and comments. I've tried to address most of them and welcome more feedback.
Hello @lheagy and @choldgraf, I welcome more feedback when you get a chance. Thanks!
I took another pass through the README - I think that it is definitely improved!
I still have a few issues that I'd like to see addressed, particularly around clarifying the inputs/outputs of the script, on improving the test coverage, and on clarifying how you can incorporate this into a full GUI visualization.
I've added comments and questions to each of the issues from before
Thanks. I've tried to address your most recent comments.
Please let us know if there are other issues that need addressed @lheagy and @choldgraf .
👋 Hi @choldgraf, would you be willing to take another look? If so, do you have an estimate of when you might be able to? Thanks!
hey - a little slammed right now, but will try to get to it ASAP. My slammed-ness will continue through the end of next week, after which it'll calm down. So if I find a moment to take a look at this before then, I will, but otherwise it'll be after the end of next week.
Thanks @choldgraf for the update. Best of luck getting through the slammed-ness!!
Thank you both.
:wave: Hi @choldgraf: a friendly reminder :) Whenever you have a chance, would you mind taking another look? Thanks!
I took a look at the latest changes to the repository, and I think that it's much improved. There's more information in there now about the reasoning behind the package, and how to use it. I consolidated some of the material that was in the python script header as well as the JOSS paper and placed it in the README in order to make it discoverable (https://github.com/rheiland/xml2jupyter/pull/6). I think that beyond this, my comments have been addressed!
Thank you both! I've merged Chris's PR and replied to his suggestion on looking into pytest.
Excellent, many thanks @choldgraf for your comments and nice work addressing these comments @rheiland! @choldgraf: there are a few unchecked items on the checklist above - if those have all been addressed, would you mind ticking them off, or if there are items that still need to be taken care of, please let us know. Thank you both!
a couple quick questions:
- It asks to list authors, there is the primary author in the metadata of the paper.md file, so I'm assuming that's a ✔️
- regarding the version, this repository isn't installable via a package manager, it's "just" a python script that is run at the command line, so I'm not sure how to answer this one. There is a release made, so I'm just considering this a ✔️ as well
- For the references, I believe that the DOI is missing for Thomas Kluyver's article: (doi:10.3233/978-1-61499-649-1-87) as well as for the scipy reference (DOI:10.1109/MCSE.2007.58)
@choldgraf, regarding the scipy ref DOI, the DOI you provided is for "Python for Scientific Computing" by Oliphant. Are you saying that's the preferred ref for citing scipy? If so, I'll need to change the entire bib entry. I was trying to follow advice here: https://www.scipy.org/citing.html
Ah, I was just copy pasting the DOIs that I had found next to the references on the scipy docs. But if you've thought it through then I am +1 on whatever you came up with
Giving this a bit more consideration, I decided to use the Oliphant 2007 citation. SciPy requires NumPy after all, which is covered in the 2007 article; it also gives deserved credit to Oliphant. Thanks for your attention to detail on these DOIs.
@whedon check references
Attempting to check references...
```Reference check summary:
OK DOIs
- 10.1371/journal.pcbi.1005991 is OK
- 10.1101/090696 is OK
- 10.1109/MCSE.2007.55 is OK
- 10.3233/978-1-61499-649-1-87 is OK
- 10.1515/ntrev-2012-0043 is OK
- 10.1038/d41586-018-07196-1 is OK
- 10.1109/MCSE.2007.58 is OK
MISSING DOIs
- None
INVALID DOIs
- None
```
@whedon add @lheagy as reviewer
OK, @lheagy is now a reviewer
:wave: @arfon (sorry for asking this again, I know you told me the solution, but I couldn't find the issue where we corrected this elsewhere!): Is there a way to keep a block of code together on one page? Is it something along the lines of a \clearpage in the paper? Thanks for your help!
I think you can do \newpage ?
:wave: @rheiland, would you like to try updating the paper so that the codeblock isn't split over two pages? (e.g. use a \newpage before to push all of the content to a new page)
@lheagy If you're saying to literally add that command to the paper.md (before the XML code block), I did so, and pushed it. But I confess I don't understand the magic you use to generate the pdf from the .md. As you can see from https://github.com/rheiland/xml2jupyter/blob/master/paper/gen_pdf.sh , I came up with my own hack to generate a .pdf, which didn't split the XML block across pages, fwiw:
https://github.com/rheiland/xml2jupyter/blob/master/paper/paper.pdf
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@rheiland you can run @whedon generate pdf yourself too :-) The command we're running is a bit of a monster: https://github.com/openjournals/whedon/blob/c34e1031c081b4e24cd7141023ed81b643c04364/lib/whedon/compilers.rb#L126-L149
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@arfon Thanks for pointing out the fact that I could have whedon generate the pdf myself (apologies for not remembering that when I listed whedon's commands a long time ago).
Some things that are less than ideal in the pdf, but not sure what, if anything, can be done about them. And besides, they are not critical.
have Figure 1 appear before the associated XML block (and the paragraph that mentions it).
have a filename be split across lines in the pdf:
The xml2jupyter.py script parses the XML and generates a Python module, user_param
s.py, ...
- have Figure 1 appear before the associated XML block (and the paragraph that mentions it).
Just to confirm, do you want the XML before the figure or vice versa? The current proof looks like this to me:

The XML before the figure would be more logical.
On Tue, Jun 25, 2019 at 8:04 AM Arfon Smith notifications@github.com
wrote:
>
- have Figure 1 appear before the associated XML block (and the
paragraph that mentions it).Just to confirm, do you want the XML before the figure or vice versa? The
current proof looks like this to me:[image: Screen Shot 2019-06-25 at 8 03 29 AM]
https://user-images.githubusercontent.com/4483/60096768-cff17900-971f-11e9-98d4-0a231868f18e.png—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
https://github.com/openjournals/joss-reviews/issues/1408?email_source=notifications&email_token=AANRKRIUA77P7WLJ2OMYZH3P4ICUBA5CNFSM4HH7EUAKYY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGODYQAGRA#issuecomment-505414468,
or mute the thread
https://github.com/notifications/unsubscribe-auth/AANRKRMJHBNMKV4ITTN7AMLP4ICUBANCNFSM4HH7EUAA
.
@whedon generate pdf from branch arfon:patch-1
Attempting PDF compilation from custom branch arfon:patch-1. Reticulating splines etc...
@rheiland - I don't think Whedon knows how to build a change from my pull request (https://github.com/rheiland/xml2jupyter/pull/7) but when I build the PDF locally with the changes in https://github.com/rheiland/xml2jupyter/pull/7 I get this:
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
How's that looking @rheiland?
Beautiful, IMO :-) Thanks for your PR! In fact, the pdf now looks very
similar to the one I generated using my hacky script.
On Tue, Jun 25, 2019 at 11:28 AM Arfon Smith notifications@github.com
wrote:
How's that looking @rheiland https://github.com/rheiland?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
https://github.com/openjournals/joss-reviews/issues/1408?email_source=notifications&email_token=AANRKRI5DYCG3I7FLSE7OP3P4I2TRA5CNFSM4HH7EUAKYY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGODYQUJWI#issuecomment-505496793,
or mute the thread
https://github.com/notifications/unsubscribe-auth/AANRKROVYYYWDQ2YLVJL7UDP4I2TRANCNFSM4HH7EUAA
.
Excellent, thanks for your help @arfon 🎉
@rheiland: can you please generate a new release of xml2jupyter, archive it on zenodo or similar and post the doi here. Please make sure that the author list and title match the those on the paper.
@lheagy: will it be OK to leave my gen_pdf.sh script (and a resulting .pdf) in the /paper directory when I generate a new release, or would that cause problems/confusion?
I also have a /preprint directory that I will remove before doing a new release.
@rheiland: I think leaving the gen_pdf.sh is fine, but the pdf might cause confusion (particularly as the DOI is not assigned in the one in your repo). So I would recommend removing it. If you wanted to keep this in your repo, then keeping the preprint directory and moving the pdf there would be appropriate.
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
:wave: @rheiland, please ping when you have had a chance to archive the software and we can proceed with publication. Thanks!
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
@lheagy @arfon if there's anything that can be done to prevent the line break of a filename (below) in the pdf generation, I'd welcome it. Otherwise, we'll move ahead with generating a release and getting a DOI. Thanks!
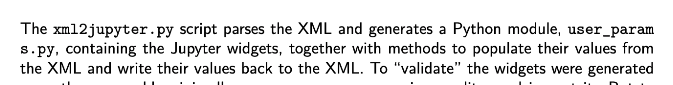

@lheagy @arfon if there's anything that can be done to prevent the line break of a filename (below) in the pdf generation, I'd welcome it. Otherwise, we'll move ahead with generating a release and getting a DOI. Thanks!
I'm afraid not. The only thing I can suggest at this point is to slightly adjust the language (lengthen or shorten) to try and force a formatting change.
@arfon OK, no worries. I did play a bit with wording, but still ended up with name split across lines, so let's go with what's there.
@lheagy I'm still new to this Zenodo-GitHub-DOI workflow, so I'd welcome feedback if I've mangled it. I did get a DOI that supposedly goes with the (JOSS) Release of our repo - it's 10.5281/zenodo.3261541 .
Is there more I need to do?
Thanks so much to @arfon and @lheagy for all your help in guiding us through the review process! We're okay to let the line break issue go: very much at the point of rapidly diminishing returns, and the proof looks fantastic otherwise!
I noticed an unchecked box on references, but so far as I can tell, all references have a DOI. Is there something else we should fix there?
Thank you! - Paul
@whedon check references
Attempting to check references...
```Reference check summary:
OK DOIs
- 10.1371/journal.pcbi.1005991 is OK
- 10.1101/090696 is OK
- 10.1109/MCSE.2007.55 is OK
- 10.3233/978-1-61499-649-1-87 is OK
- 10.1515/ntrev-2012-0043 is OK
- 10.1038/d41586-018-07196-1 is OK
- 10.1109/MCSE.2007.58 is OK
MISSING DOIs
- None
INVALID DOIs
- None
```
At this point I think we're just waiting for the final 👍from @lheagy and then I think we're good to accept here.
Fantastic! Thank you!
@whedon set 10.5281/zenodo.3261541 as archive
OK. 10.5281/zenodo.3261541 is the archive.
:wave: @rheiland: would you mind updating the title of the Zenodo archive to be: "xml2jupyter: Mapping parameters between XML and Jupyter widgets" (the same as your paper title)? Please also add all of the authors that are listed on the paper to the archive record: Randy Heiland, Daniel Mishler, Tyler Zhang, Eric Bower, and Paul Macklin.
Thanks!
Ah, thank you for continuing to educate me on this @lheagy ! Hopefully it's correct now.
@whedon accept
Attempting dry run of processing paper acceptance...
```Reference check summary:
OK DOIs
- 10.1371/journal.pcbi.1005991 is OK
- 10.1101/090696 is OK
- 10.1109/MCSE.2007.55 is OK
- 10.3233/978-1-61499-649-1-87 is OK
- 10.1515/ntrev-2012-0043 is OK
- 10.1038/d41586-018-07196-1 is OK
- 10.1109/MCSE.2007.58 is OK
MISSING DOIs
- None
INVALID DOIs
- None
```
Check final proof :point_right: https://github.com/openjournals/joss-papers/pull/806
If the paper PDF and Crossref deposit XML look good in https://github.com/openjournals/joss-papers/pull/806, then you can now move forward with accepting the submission by compiling again with the flag deposit=true e.g.
@whedon accept deposit=true
@whedon accept deposit=true
Doing it live! Attempting automated processing of paper acceptance...
🐦🐦🐦 👉 Tweet for this paper 👈 🐦🐦🐦
🚨🚨🚨 THIS IS NOT A DRILL, YOU HAVE JUST ACCEPTED A PAPER INTO JOSS! 🚨🚨🚨
Here's what you must now do:
- Check final PDF and Crossref metadata that was deposited :point_right: https://github.com/openjournals/joss-papers/pull/807
- Wait a couple of minutes to verify that the paper DOI resolves https://doi.org/10.21105/joss.01408
- If everything looks good, then close this review issue.
Party like you just published a paper! 🎉🌈🦄💃👻🤘
Any issues? notify your editorial technical team...
@choldgraf, @lheagy - many thanks for your reviews here and to @lheagy for editing this submission ✨
@rheiland - your paper is now accepted into JOSS :zap::rocket::boom:
:tada::tada::tada: Congratulations on your paper acceptance! :tada::tada::tada:
If you would like to include a link to your paper from your README use the following code snippets:
Markdown:
[](https://doi.org/10.21105/joss.01408)
HTML:
<a style="border-width:0" href="https://doi.org/10.21105/joss.01408">
<img src="http://joss.theoj.org/papers/10.21105/joss.01408/status.svg" alt="DOI badge" >
</a>
reStructuredText:
.. image:: http://joss.theoj.org/papers/10.21105/joss.01408/status.svg
:target: https://doi.org/10.21105/joss.01408
This is how it will look in your documentation:
We need your help!
Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider doing either one (or both) of the the following:
- Volunteering to review for us sometime in the future. You can add your name to the reviewer list here: http://joss.theoj.org/reviewer-signup.html
- Making a small donation to support our running costs here: https://numfocus.salsalabs.org/donate-to-joss
Thank you very much @arfon, @lheagy, and esp @choldgraf for this terrific experience!
Randy
Thanks very much for leading us through our first JOSS submission! Thanks
to @rheiland for terrific work as lead author!
In case it helps your statistics for impact, etc., please note that this
paper has three undergraduate co-authors.
All the best -- Paul
>
please note that this paper has three undergraduate co-authors.
✨
please note that this paper has three undergraduate co-authors.
✨++
Most helpful comment
Thank you very much @arfon, @lheagy, and esp @choldgraf for this terrific experience!
Randy