Joss-reviews: [REVIEW]: Vizualizing DNA sequence co-linearity with xmatchview
Submitting author: @warrenlr (Rene L. Warren)
Repository: https://github.com/warrenlr/xmatchview
Version: v0.3
Editor: @mgymrek
Reviewer: @nmmsv
Archive: 10.5281/zenodo.1155709
Status
Status badge code:
HTML: <a href="https://joss.theoj.org/papers/002324a31f8d2fc5101f09ef52fc2777"><img src="https://joss.theoj.org/papers/002324a31f8d2fc5101f09ef52fc2777/status.svg"></a>
Markdown: [](https://joss.theoj.org/papers/002324a31f8d2fc5101f09ef52fc2777)
Reviewers and authors:
Please avoid lengthy details of difficulties in the review thread. Instead, please create a new issue in the target repository and link to those issues (especially acceptance-blockers) in the review thread below. (For completists: if the target issue tracker is also on GitHub, linking the review thread in the issue or vice versa will create corresponding breadcrumb trails in the link target.)
Reviewer instructions & questions
@nmmsv, please carry out your review in this issue by updating the checklist below. If you cannot edit the checklist please:
- Make sure you're logged in to your GitHub account
- Be sure to accept the invite at this URL: https://github.com/openjournals/joss-reviews/invitations
The reviewer guidelines are available here: https://joss.theoj.org/about#reviewer_guidelines. Any questions/concerns please let @mgymrek know.
Conflict of interest
- [x] As the reviewer I confirm that I have read the JOSS conflict of interest policy and that there are no conflicts of interest for me to review this work.
Code of Conduct
- [x] I confirm that I read and will adhere to the JOSS code of conduct.
General checks
- [x] Repository: Is the source code for this software available at the repository url?
- [x] License: Does the repository contain a plain-text LICENSE file with the contents of an OSI approved software license?
- [x] Version: Does the release version given match the GitHub release (v0.3)?
- [x] Authorship: Has the submitting author (@warrenlr) made major contributions to the software? Does the full list of paper authors seem appropriate and complete?
Functionality
- [x] Installation: Does installation proceed as outlined in the documentation?
- [x] Functionality: Have the functional claims of the software been confirmed?
- [x] Performance: If there are any performance claims of the software, have they been confirmed? (If there are no claims, please check off this item.)
Documentation
- [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] Installation instructions: Is there a clearly-stated list of dependencies? Ideally these should be handled with an automated package management solution.
- [x] Example usage: Do the authors include examples of how to use the software (ideally to solve real-world analysis problems).
- [x] Functionality documentation: Is the core functionality of the software documented to a satisfactory level (e.g., API method documentation)?
- [x] Automated tests: Are there automated tests or manual steps described so that the function of the software can be verified?
- [x] Community guidelines: Are there clear guidelines for third parties wishing to 1) Contribute to the software 2) Report issues or problems with the software 3) Seek support
Software paper
- [x] Authors: Does the
paper.mdfile include a list of authors with their affiliations? - [x] A statement of need: Do the authors clearly state what problems the software is designed to solve and who the target audience is?
- [x] References: Do all archival references that should have a DOI list one (e.g., papers, datasets, software)?
All 40 comments
Hello human, I'm @whedon. I'm here to help you with some common editorial tasks. @nmmsv it looks like you're currently assigned as the reviewer for this paper :tada:.
:star: Important :star:
If you haven't already, you should seriously consider unsubscribing from GitHub notifications for this (https://github.com/openjournals/joss-reviews) repository. As a reviewer, you're probably currently watching this repository which means for GitHub's default behaviour you will receive notifications (emails) for all reviews 😿
To fix this do the following two things:
- Set yourself as 'Not watching' https://github.com/openjournals/joss-reviews:

- You may also like to change your default settings for this watching repositories in your GitHub profile here: https://github.com/settings/notifications

For a list of things I can do to help you, just type:
@whedon commands
Attempting PDF compilation. Reticulating splines etc...
PDF failed to compile for issue #497 with the following error:
Killed
Looks like we failed to compile the PDF
Thanks @nmmsv for agreeing to review!
@warrenlr @arfon it looks like there are some issues compiling the PDF. It sounds like this issue was discussed in the pre-review thread. Was this resolved?
not to my knowledge. I believe it has to be compiled manually on a local machine (Fig. 3 is rather large). The latest and current build is here:
https://github.com/openjournals/joss-reviews/files/1548851/10.21105.joss.00491.pdf
thanks!
Rene
@warrenlr @arfon it looks like there are some issues compiling the PDF. It sounds like this issue was discussed in the pre-review thread. Was this resolved?
It was not. The PDF is too big for our current approach so I had to manually produce it as @warrenlr said. The URL he shared above is the latest one.
thank you for agreeing to review, @nmmsv! Were you able to access the pdf of the submitted manuscript? Thanks, Rene
Hi @warrenlr , absolutely! I was able to access the pdf file.
I had some issues trying to install the software. I list them here:
It is required to use python 2.3 or 2.4. These releases are quite outdated (latest release from 2008) and some operating systems may not support them. I managed to find a compatible version for our linux system.
It is required to use windows font ttf files, which are not accessible to me (and I assume others that don't have a windows system around). Also, I'm not sure if it's possible to use windows fonts under GNU General Public license.
Another dependency is cross_match. My understanding is that this software is not open source. In order to use it (even for academic purposes), users need to complete a user agreement and send it alongside a unique ID to 3 different emails to get permission to use.
http://www.phrap.org/consed/consed.html#howToGetIn the dependencies sections, there are ambiguous mentions to some lines of the codes that need to be manually modified, as well as ambiguous words (i.e., "below").
Please let me know if you had any questions about my comments.
Best,
Nima
Thank you for your comments, @nmmsv.
What do you advise I do going forward?
In the mean time, I clarified the dependencies in the README.md:
You will need to do the following before you can proceed:
1) Download python2.3 or 2.4 from: http://www.python.org/ (The code may work with newer versions of python (not tested))
2) Download the Python Imaging Library (PIL) from: http://www.pythonware.com/products/pil/
3) Obtain true type fonts (ttf) and change the corresponding lines in xmatchview.py and xmatchview-conifer.py to reflect the location of the ttf. Alternatively, you could use fonts supplied by PIL instead (eg. ./pil/helvB12.pil)
4) Download cross_match for academic use, see http://www.phrap.org and http://www.phrap.org/consed/consed.html#howToGet
5) Make sure cross_match is in your $PATH
Here are some of my comments:
- I haven't updated/tested the code with newer python releases.
Since I am not distributing the fonts with xmatchview, the code itself is still GPL, I think.
Beside, arial ttf fonts are available for linux:
eg.
https://github.com/JotJunior/PHP-Boleto-ZF2/tree/master/public/assets/fonts
Also, the code may be modified to use fonts that come with PIL (eg. ./pil/helvB12.pil)Yes, xmv depends on cross_match as stated in the documentation/paper. It is unfortunate that cross_match is not easier to obtain, but out of my control.
I have deleted these old requirements, they were for an older version of the code.
Let me know how to proceed from here.
Thank you!
Rene
@warrenlr
Please create input options for font or any other part that is expected to be modified in source code. This way users can supply the font path (and other options) as input.
I also requested a cross_match license. They say that it may take up to 2 weeks, so I'll be waiting to hear from them.
Best,
Nima
Thanks for the clarifications.
echoing @nmmsv: Just scrolling through the source code and based on comments of @nmmsv, regarding fonts I see many of the paths are hard coded in the main .py file and need to be changed before running. It would be better to make these have reasonable defaults and allow paths to be changed via a command line argument. Also more info on how to download the required fonts, or if possible distributing those with the package, would make installation easier.
thank you @mgymrek and @nmmsv for the helpful comments and diligence.
I have streamlined the code and added the suggested option to provide a path to the font directory. I added some information to the README.md regarding font usage.
850e689959597cbadbada4e4ef96b05af0b2aeb2
b902c85b96afba2eab55e0786c7830c5e7d1c686
fyi - I have added a test folder with FASTA and cross_match output files for one of the FTL1 example from Fig.3. Users can now test their install (does not require cross_match).
Thank you
Rene
Hello,
I did receive the license for cross_match. But I'll be trying to run the test script for now. There are some modifications that help the user in running the code, and also an error log that I've listed below.
Please make test script customizable, ask for inputs instead of setting manual paths (Also create usage and --help input to explain the requirements). For example, in runXMV.sh there are python paths to folders in your personal computer's home directory. Please explain (in usage or help) which paths should be used instead of those and provide a proper input mechanism.
Please avoid having a separate copy of xmatchview.py for the test folder. Right now there seems to be two separate source codes which creates confusion when edits are made.
After some path modifications and changes, I could run runXMV.sh, and there seems to be a problem when using Image library. I've attached the log and error of the program below.
Thank you.
Nima
thank you.
- done.
- created symbolic links
- I tracked this problem (I am not seeing this on my system) and fixed according to instructions here: https://stackoverflow.com/questions/17451711/typeerror-when-resizing-an-image-with-pil-in-python
Let me know if this fixes it. If not, may have to change all calls to PIL modules (eg. from PIL import XX)
thanks
Rene
Hello,
I now realize that my previous log was truncated for some reason and didn't contain the actual error message. I tried again with the new version of the code, and I still get the following error:
2629 out of 2635
2630 out of 2635
2631 out of 2635
2632 out of 2635
2633 out of 2635
2634 out of 2635
done.
Drawing repeats...
Traceback (most recent call last):
File "xmatchview.py", line 800, in
main()
File "xmatchview.py", line 795, in main
drawRelationship(reference, query, match, scale, query_hit, mismatch, block_length, crossmatch_file, freq, reflength, leap, format, formatdict, protein, alpha, refexon, qryexon, qrylength, refnpos, qrynpos, refname, qryname, refzpos, qryzpos, fontpath)
File "xmatchview.py", line 420, in drawRelationship
bdraw = ImageDraw.Draw(back)
File "/storage/nmmsv/xmatchview/Imaging-1.1.7/PIL/ImageDraw.py", line 294, in Draw
return ImageDraw(im, mode)
File "/storage/nmmsv/xmatchview/Imaging-1.1.7/PIL/ImageDraw.py", line 75, in __init__
self.draw = Image.core.draw(self.im, blend)
TypeError: must be ImagingCore, not ImagingCore
Apologies for confusing log.
Best,
Nima
Hi Nima,
I have preceded each PIL module import statement by "from PIL"
hopefully this fixes it.
Thanks,
Rene
Hello @nmmsv, has the fix worked on your system?
Hello @warrenlr ,
Sorry I was busy yesterday. believe now things are running as they should. I've attached the resulting figure.
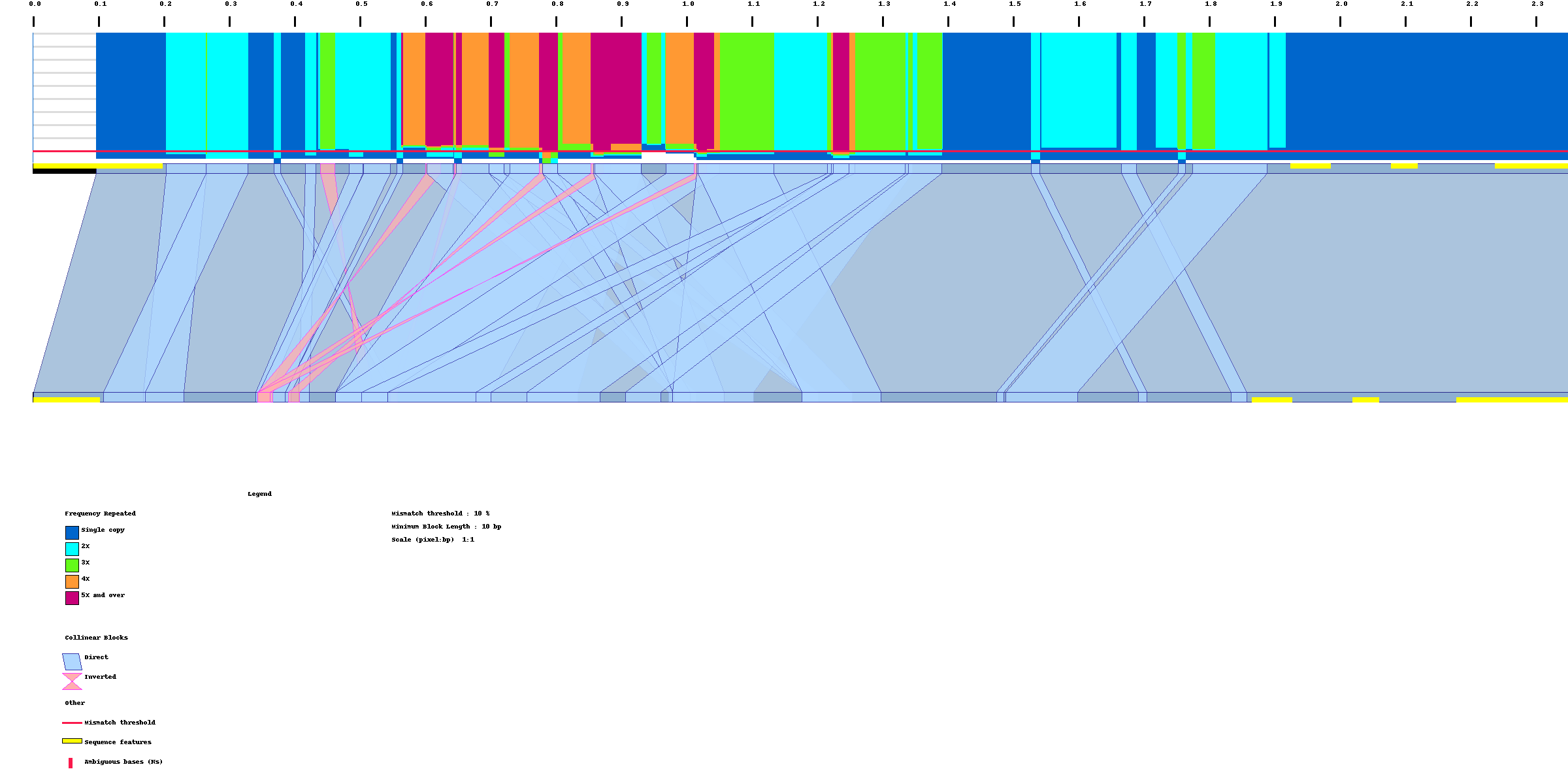
I'll go ahead and read the documentation and let you know if I had questions.
Best,
Nima
Hi @warrenlr ,
I've read your paper, and I have a few comments based on the reviewer guidelines:
- Please remove API explanation from paper, and add them to the documentation.
- Test: Please create a means for the user to test proper installation and correct functionality. Ideally, we want to make sure the user can test every option and make sure they all work. Currently, there are example scripts in the test folder (which are another required item), but the user cannot confirm the results and correct functionality.
Thanks.
Best,
Nima
thank you @nmmsv
Could you confirm that the sentence you wish to remove from the paper and included in the readme is:
"Demo shell scripts that pipeline cross_match and xmatchview/xmatchview-conifer are included with the distribution and provided for guidance (runCompareTwoGenomesColinear.sh and runSpruceView.sh, respectively)" ?I am not sure I understand what you require here, that is not already provided. For this, I have created a test directory where users can test their install by comparing the figure they generate with the ones provided. In the readme, I have a special section where I indicate:
TEST xmatchview.py / xmatchview-conifer.py
At the unix prompt, once the package is installed, change directory to
cd ./test
Once you have downloaded pyhon and PIL and changed the paths to fonts in the xmatchview.py and xmatchview-conifer.py Execute:
./runXMV-conifer.sh FTL1_ss.fa_vs_FTL1_pa.fa.rep FTL1_ss.fa FTL1_pa.fa 200 10 1 FTL1_ss.txt FTL1_pa.txt
and
./runXMV.sh FTL1_ss.fa_vs_FTL1_pa.fa.rep FTL1_ss.fa FTL1_pa.fa 200 10 1 FTL1_ss.txt FTL1_pa.txt
If all went well, images such as those provided in the test folder should be generated
they can confirm results by comparing the resulting images. The functionality / parameters can be tested on these examples if need be.
Final comment:
At this point, with your insightful comments (which I am grateful for), xmatchview has been tested and demonstrated to work in your hands. I have also updated the readme since the original submission to clarify dependencies, installation and usability, thanks to your comments and that of the Editor, @mgymrek. The manuscript describes the utility of the tool, and with supporting citations, presents use cases. I trust that this is now sufficient, unless there are specifics you wish to see addressed, edited, removed, or added. Regards, Rene
I made the requested changes. Please see commit:
https://github.com/warrenlr/xmatchview/commit/3e1f5eb4db02a9ffc90763a252a168e15dc0ba4c
Hi @warrenlr ,
- I was referring to the part that you explain in detail how the software works and each input feature and flag. Based on the guidelines, I think the paper should be more of an overview of motivation, high level functionality, and some applications:
> Note the paper should not include software documentation such as API (Application Programming Interface) functionality, as this should be outlined in the software documentation. - Thanks, the test procedure is now more clear with your explanation in the documentation.
Best,
Nima
Hello @nmmsv ,
- got it. I have made the recommended changes in the latest commit.
https://github.com/warrenlr/xmatchview/commit/833529e64e655826f785136830e8734eea18da91
Thanks,
Rene
Hi @warrenlr ,
Thanks for the updates.
I currently can't see the references in the paper.md file, I think they should be in the bib file? Can you please add the final version of the paper (pdf) to the repository? I think that version should contain everything.
Best,
Nima
@whedon generate pdf
Here are some things you can ask me to do:
# List all of Whedon's capabilities
@whedon commands
# Assign a GitHub user as the reviewer of this submission
@whedon assign @username as reviewer
# List of editor GitHub usernames
@whedon list editors
# List of reviewers together with programming language preferences and domain expertise
@whedon list reviewers
# Change editorial assignment
@whedon assign @username as editor
# Set the software archive DOI at the top of the issue e.g.
@whedon set 10.0000/zenodo.00000 as archive
# Open the review issue
@whedon start review
🚧 🚧 🚧 Experimental Whedon features 🚧 🚧 🚧
# Compile the paper
@whedon generate pdf
Trying to re-generate the pdf, hopefully won't crash the server.. @arfon , just looping you in..may need your assistance.
@whedon generate pdf
Attempting PDF compilation. Reticulating splines etc...
Here's the latest compiled PDF: 10.21105.joss.00497.pdf
thank you @arfon
hi @nmmsv , I added the latest compiled version of the paper (pdf) to the repository.
thank you
Rene
Hi @warrenlr ,
Perfect, I believe all the criteria has been met!
@mgymrek mentioning you to let you know and ask if any further actions needs to be taken.
Best,
Nima
awesome, thank you for your review @nmmsv !
thanks @nmmsv and @warrenlr!
@arfon, I think we're ready to accept.
@warrenlr - At this point could you make an archive of the reviewed software in Zenodo/figshare/other service and update this thread with the DOI of the archive? I can then move forward with accepting the submission.
@arfon, I have archived the software on zenodo 10.5281/zenodo.1155709
@whedon set 10.5281/zenodo.1155709 as archive
OK. 10.5281/zenodo.1155709 is the archive.
@nmmsv - many thanks for your review here and to @mgymrek for editing this one ✨
@warrenlr - your paper is now accepted into JOSS and your DOI is https://doi.org/10.21105/joss.00497 ⚡️ 🚀 💥
:tada::tada::tada: Congratulations on your paper acceptance! :tada::tada::tada:
If you would like to include a link to your paper from your README use the following code snippet:
[](https://doi.org/10.21105/joss.00497)
This is how it will look in your documentation:
We need your help!
Journal of Open Source Software is a community-run journal and relies upon volunteer effort. If you'd like to support us please consider volunteering to review for us sometime in the future. You can add your name to the reviewer list here: http://joss.theoj.org/reviewer-signup.html
Most helpful comment
@nmmsv - many thanks for your review here and to @mgymrek for editing this one ✨
@warrenlr - your paper is now accepted into JOSS and your DOI is https://doi.org/10.21105/joss.00497 ⚡️ 🚀 💥