Dplyr: dbplyr + odbc Access connections use TOP not LIMIT - tbl won't print
Issue Description and Expected Result
I believe that this issue is similar to Microsoft SQL Server, but currently tbl() fails on creating a usable connection to an Access database table. The connection to the database was made through odbc.
More specifically, the sql generated from print.tbl_sql() is incorrect because Access does not support LIMIT and instead uses TOP.
I think the correct approach is to use the sql_select.Microsoft SQL Server() that already exists.
Reprex
I understand that you cannot fully reproduce this without an Access database included, but hopefully you can run with it.
Notice the "Syntax error in FROM clause".
Connect to Access and create tbl
library(DBI)
library(dbplyr)
suppressPackageStartupMessages(library(dplyr))
#> Warning: package 'dplyr' was built under R version 3.4.1
# Connecting (works and gives warning, already discussed with Jim Hester)
cn <- dbConnect(odbc::odbc(), dsn = "access-odbc")
#> Warning message:
#> Could not notify connection observer. nanodbc/nanodbc.cpp:4274: HYC00: [Microsoft][ODBC Microsoft Access Driver]Optional feature not implemented
# Create a simple table and add data
# dbGetQuery(cn, "DROP TABLE testing")
dbGetQuery(cn, "CREATE TABLE testing (Col1 VarChar, Col2 Double)")
#> data frame with 0 columns and 0 rows
dbGetQuery(cn, "INSERT INTO testing (Col1, Col2) VALUES ('hello', 2)")
#> data frame with 0 columns and 0 rows
test_table <- dbGetQuery(cn, "SELECT * FROM testing")
test_table
#> Col1 Col2
#> 1 hello 2
# Create a tbl object from that table and print it, no good
test_tbl <- tbl(cn, "testing")
test_tbl
#> Error in new_result(connection@ptr, statement): nanodbc/nanodbc.cpp:1344: 42000: [Microsoft][ODBC Microsoft Access Driver] Syntax error in FROM clause.
My solution
Use a custom sql_select.ACCESS(). It's the same as Microsoft SQL Server.
library(DBI)
library(dbplyr)
suppressPackageStartupMessages(library(dplyr))
#> Warning: package 'dplyr' was built under R version 3.4.1
cn <- dbConnect(odbc::odbc(), dsn = "access-odbc")
### SQL SELECT FOR ACCESS - Copied from Microsoft SQL Server --------------------------------------
`sql_select.ACCESS`<- function(con, select, from, where = NULL,
group_by = NULL, having = NULL,
order_by = NULL,
limit = NULL,
distinct = FALSE,
...) {
out <- vector("list", 7)
names(out) <- c("select", "from", "where", "group_by",
"having", "order_by","limit")
assertthat::assert_that(is.character(select), length(select) > 0L)
out$select <- build_sql(
"SELECT ",
if (distinct) sql("DISTINCT "),
# MS SQL uses the TOP statement instead of LIMIT which is what SQL92 uses
# TOP is expected after DISTINCT and not at the end of the query
# e.g: SELECT TOP 100 * FROM my_table
if (!is.null(limit) && !identical(limit, Inf)) {
assertthat::assert_that(is.numeric(limit), length(limit) == 1L, limit > 0)
build_sql(" TOP ", as.integer(limit), " ")},
escape(select, collapse = ", ", con = con)
)
out$from <- sql_clause_from(from, con)
out$where <- sql_clause_where(where, con)
out$group_by <- sql_clause_group_by(group_by, con)
out$having <- sql_clause_having(having, con)
out$order_by <- sql_clause_order_by(order_by, con)
escape(unname(dplyr:::compact(out)), collapse = "\n", parens = FALSE, con = con)
}
### Helpers needed to run this ---------------------------------------------------------------
sql_clause_generic <- function(clause, fields, con){
if (length(fields) > 0L) {
assert_that(is.character(fields))
build_sql(
sql(clause), " ",
escape(fields, collapse = ", ", con = con)
)
}
}
sql_clause_select <- function(select, con, distinct = FALSE){
assert_that(is.character(select))
if (is_empty(select)) {
abort("Query contains no columns")
}
build_sql(
"SELECT ",
if (distinct) sql("DISTINCT "),
escape(select, collapse = ", ", con = con)
)
}
sql_clause_where <- function(where, con){
if (length(where) > 0L) {
assert_that(is.character(where))
where_paren <- escape(where, parens = TRUE, con = con)
build_sql("WHERE ", sql_vector(where_paren, collapse = " AND "))
}
}
sql_clause_limit <- function(limit, con){
if (!is.null(limit) && !identical(limit, Inf)) {
assert_that(is.numeric(limit), length(limit) == 1L, limit >= 0)
build_sql(
"LIMIT ", sql(format(trunc(limit), scientific = FALSE)),
con = con
)
}
}
sql_clause_from <- function(from, con) sql_clause_generic("FROM", from, con)
sql_clause_group_by <- function(group_by, con) sql_clause_generic("GROUP BY", group_by, con)
sql_clause_having <- function(having, con) sql_clause_generic("HAVING", having, con)
sql_clause_order_by <- function(order_by, con) sql_clause_generic("ORDER BY", order_by, con)
### Now it works! --------------------------------------------------------------------------------------
test_tbl <- tbl(cn, "testing")
test_tbl
#> # Source: table<testing> [?? x 2]
#> # Database: ACCESS 12.00.0000[admin@ACCESS/R:\Life_Modeling_Team\Vaughan\Economic Scenario Database 2017 Part 1.accdb]
#> Col1 Col2
#> <chr> <dbl>
#> 1 hello 2
Other details
I am on macOS Sierra. Running R 3.4.0
Database info
library(DBI)
cn <- dbConnect(odbc::odbc(), dsn = "access-odbc")
dbGetInfo(cn)
#> $dbname
#> [1] "R:\\Life_Modeling_Team\\Vaughan\\Economic Scenario Database 2017 Part 1.accdb"
#>
#> $dbms.name
#> [1] "ACCESS"
#>
#> $db.version
#> [1] "12.00.0000"
#>
#> $username
#> [1] "admin"
#>
#> $host
#> [1] ""
#>
#> $port
#> [1] ""
#>
#> $sourcename
#> [1] "access-odbc"
#>
#> $servername
#> [1] "ACCESS"
#>
#> $drivername
#> [1] "ACEODBC.DLL"
#>
#> $odbc.version
#> [1] "03.80.0000"
#>
#> $driver.version
#> [1] "Microsoft Access database engine"
#>
#> $odbcdriver.version
#> [1] "03.51"
#>
#> $supports.transactions
#> [1] FALSE
#>
#> attr(,"class")
#> [1] "ACCESS" "driver_info" "list"
Session info
Session Info
library(DBI)
library(dbplyr)
library(odbc)
#> Warning: package 'odbc' was built under R version 3.4.1
library(dplyr)
#> Warning: package 'dplyr' was built under R version 3.4.1
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:dbplyr':
#>
#> ident, sql
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
devtools::session_info()
#> Session info -------------------------------------------------------------
#> setting value
#> version R version 3.4.0 (2017-04-21)
#> system i386, mingw32
#> ui RTerm
#> language (EN)
#> collate English_United States.1252
#> tz America/New_York
#> date 2017-07-06
#> Packages -----------------------------------------------------------------
#> package * version date source
#> assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
#> backports 1.1.0 2017-05-22 CRAN (R 3.4.0)
#> base * 3.4.0 2017-04-21 local
#> bindr 0.1 2016-11-13 CRAN (R 3.4.0)
#> bindrcpp 0.2 2017-06-17 CRAN (R 3.4.0)
#> bit 1.1-12 2014-04-09 CRAN (R 3.4.0)
#> bit64 0.9-7 2017-05-08 CRAN (R 3.4.0)
#> blob 1.1.0 2017-06-21 Github (hadley/blob@9dd54d9)
#> compiler 3.4.0 2017-04-21 local
#> datasets * 3.4.0 2017-04-21 local
#> DBI * 0.7 2017-06-18 CRAN (R 3.4.0)
#> dbplyr * 1.0.0 2017-06-09 CRAN (R 3.4.0)
#> devtools 1.13.2 2017-06-02 CRAN (R 3.4.0)
#> digest 0.6.12 2017-01-27 CRAN (R 3.4.0)
#> dplyr * 0.7.1 2017-06-22 CRAN (R 3.4.1)
#> evaluate 0.10.1 2017-06-24 CRAN (R 3.4.0)
#> glue 1.1.0 2017-06-13 CRAN (R 3.4.0)
#> graphics * 3.4.0 2017-04-21 local
#> grDevices * 3.4.0 2017-04-21 local
#> hms 0.3 2016-11-22 CRAN (R 3.4.0)
#> htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
#> knitr 1.16 2017-05-18 CRAN (R 3.4.0)
#> magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
#> memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
#> methods * 3.4.0 2017-04-21 local
#> odbc * 1.1.1 2017-06-27 CRAN (R 3.4.1)
#> pkgconfig 2.0.1 2017-03-21 CRAN (R 3.4.0)
#> R6 2.2.2 2017-06-17 CRAN (R 3.4.0)
#> Rcpp 0.12.11 2017-05-22 CRAN (R 3.4.0)
#> rlang 0.1.1.9000 2017-06-28 Github (hadley/rlang@3e4f785)
#> rmarkdown 1.6 2017-06-15 CRAN (R 3.4.0)
#> rprojroot 1.2 2017-01-16 CRAN (R 3.4.0)
#> stats * 3.4.0 2017-04-21 local
#> stringi 1.1.5 2017-04-07 CRAN (R 3.4.0)
#> stringr 1.2.0 2017-02-18 CRAN (R 3.4.0)
#> tibble 1.3.3 2017-05-28 CRAN (R 3.4.0)
#> tools 3.4.0 2017-04-21 local
#> utils * 3.4.0 2017-04-21 local
#> withr 1.0.2 2016-06-20 CRAN (R 3.4.0)
#> yaml 2.1.14 2016-11-12 CRAN (R 3.4.0)
All 47 comments
Hi @DavisVaughan , you're correct, Access uses TOP just like MSSQL, but there are enough syntax differences between the two that we really need a custom translation for Access. There's another Issue opened for Access #2925. If this is something you'd be interested in helping us with, please start here: http://db.rstudio.com/translation , I found a quick guide that outlines the differences between the TSQL and Access SQL: http://rogersaccessblog.blogspot.com/2013/05/what-are-differences-between-access-sql.html
@edgararuiz, I'm happy to offer some help to get this working. Enough corporate businesses use Access that I think it would be worthwhile to invest in this, as it would make it easier for them to access their data without needing to know SQL. I'll check out the guide and get back to you. Perhaps @dmenne and I can work together on this.
If I have questions along the way, where is the best place to ask them? Thanks!
That's great!
If you plan to start soon, I'd suggest to use this Issue for communication. We can transition to something else if it becomes necessary.
To get you started, the translation should be a new R script in dbplyr called db-odbc-access.R. I'd suggest to start with a copy of the MSSQL translation (https://github.com/tidyverse/dbplyr/blob/master/R/db-odbc-mssql.R) and modify it to fit Access' syntax.
@edgararuiz, making good progress over here. I've got working sql_select.ACCESS() and sql_translate_env.ACCESS() functions. They exist here. I've also got a file that runs a number of test queries (currently very ad hoc) and a Gist of it with results is here. There are a number of fixes that still need to be made.
I'll take them on 1 by 1, but let's start with a really annoying one.
select() doesn't actually work. On line 111 of the Gist, you can see that the SQL generated is:
#> <SQL>
#> SELECT `Trial` AS `Trial`
#> FROM `PE2000`
Unfortunately, Access sees this as circular referencing and blows up with the error:
Circular reference caused by alias 'Trial' in query definition's SELECT list.
Bear with me as I explain my thoughts here!
From what I can tell, while trying to debug this I find that at some point I have the object of class ident that is a named vector:
"Trial"
Printing objects of this ident class results in:
<IDENT> "Trial" AS "Trial"
Digging into print.ident, and then format.ident, I find that it is calling escape(x) where x is the ident object, and escape() looks like this.
function(x, parens = FALSE, collapse = ", ", con = NULL) {
y <- sql_escape_ident(con, x)
sql_vector(names_to_as(y, names2(x), con = con), parens, collapse)
}
Finally, the culprit seems to be names_to_as:
function(x, names = names2(x), con = NULL) {
as <- ifelse(names == "", "", paste0(" AS ", sql_escape_ident(con, names)))
paste0(x, as)
}
Which returns:
names_to_as(y, names2(x), con = con)
[1] "`Trial` AS `Trial`"
Is it really necessary for names_to_as to return in this fashion? I get it if its something like:
"`Trial` * 2 AS `Trial`"
__But when its the same name on both sides, can it just return "`Trial`"?__
For example, this extra check would work since you already return "" when there are no names:
names_to_as <- function(x, names = names2(x), con = NULL) {
as <- ifelse(names == "" | sql_escape_ident(con, names) == x, "", paste0(" AS ", sql_escape_ident(con, names)))
paste0(x, as)
}
names_to_as(y, names2(x), con = con)
[1] "`Trial`"
I changed this on my fork of dbplyr, and it seems to work great with no noticeable side effects.
Maybe I'm missing something here. Either way, some fix will have to be made since Access sees that type of aliasing as circular referencing. Thanks for the help.
Hi @DavisVaughan , great work! I'm glad you have been making progress.
I found this article in Microsoft that says that alias are possible in Access: https://support.office.com/en-us/article/Access-SQL-SELECT-clause-12d169e7-0348-407d-9c67-180ff32540ac#bm3
I have a hunch feeling that maybe changing the ident is the way to go Can you run iterate a few ways using DBI::dbGetQuery to see which quotes work (if any)?:
SELECT `Trial` AS `Trial` FROM `PE2000`
Maybe something like this:
SELECT "Trial" AS "Trial" FROM `PE2000`
or this
SELECT 'Trial' AS 'Trial' FROM `PE2000`
@edgararuiz, we are almost on the same page. It's not that aliasing as a whole doesn't work, but that aliasing _as the same name_ doesn't work, and is seen as "circular referencing."
Which means that this doesn't work:
SELECT `Trial` AS `Trial` FROM `PE2000`
But this does:
SELECT `Trial` AS `Trial2` FROM `PE2000`
The solution I suggested is simply changing the names_to_as() function to return only "`Trial`" instead of "`Trial` AS `Trial`" when the value inside the ident object is the exact same as it's name.
Example
library(DBI)
library(dbplyr)
suppressPackageStartupMessages(library(dplyr))
#> Warning: package 'dplyr' was built under R version 3.4.1
cn <- dbConnect(odbc::odbc(), dsn = "access-odbc")
# These don't work
dbGetQuery(cn, "SELECT `Trial` AS `Trial` FROM `PE2000`")
#> Error: <SQL> 'SELECT `Trial` AS `Trial` FROM `PE2000`'
#> nanodbc/nanodbc.cpp:1587: HY000: [Microsoft][ODBC Microsoft Access Driver] Circular reference caused by alias 'Trial' in query definition's SELECT list.
dbGetQuery(cn, 'SELECT "Trial" AS "Trial" FROM `PE2000`')
#> Error: <SQL> 'SELECT "Trial" AS "Trial" FROM `PE2000`'
#> nanodbc/nanodbc.cpp:1587: HY000: [Microsoft][ODBC Microsoft Access Driver] Circular reference caused by alias 'Trial' in query definition's SELECT list.
# This one isn't right
tr <- dbGetQuery(cn, "SELECT 'Trial' AS 'Trial' FROM `PE2000`")
tibble::as.tibble(tr)
#> # A tibble: 1,802,000 x 1
#> `'Trial'`
#> <chr>
#> 1 Trial
#> 2 Trial
#> 3 Trial
#> 4 Trial
#> 5 Trial
#> 6 Trial
#> 7 Trial
#> 8 Trial
#> 9 Trial
#> 10 Trial
#> # ... with 1,801,990 more rows
# This works
trial <- dbGetQuery(cn, "SELECT `Trial` AS `Trial2` FROM `PE2000`")
tibble::as.tibble(trial)
#> # A tibble: 1,802,000 x 1
#> Trial2
#> <int>
#> 1 NA
#> 2 1
#> 3 1
#> 4 1
#> 5 1
#> 6 1
#> 7 1
#> 8 1
#> 9 1
#> 10 1
#> # ... with 1,801,990 more rows
# This works
trial <- dbGetQuery(cn, "SELECT `Trial` FROM `PE2000`")
tibble::as.tibble(trial)
#> # A tibble: 1,802,000 x 1
#> Trial
#> <int>
#> 1 NA
#> 2 1
#> 3 1
#> 4 1
#> 5 1
#> 6 1
#> 7 1
#> 8 1
#> 9 1
#> 10 1
#> # ... with 1,801,990 more rows
Here is the description of the error given by Microsoft. It explains exactly what is going on. Notice how the example SELECT's A but also does an alias AS A.
https://msdn.microsoft.com/en-us/library/bb223440(v=office.12).aspx
Access doesn't like that haha.
Ok, then I suggest to apply your change to the name_to_as() function, and test your repo against other databases to make sure we didn't break anything
To get you ready for a PR, we'll need 2 more things:
- Unit tests - Please see the pattern in the MSSQL test script (https://github.com/tidyverse/dbplyr/blob/master/tests/testthat/test-translate-mssql.r) , your script should be named
test-translate-access.r - Update NEWS , please add this issue number and your GitHub name - something like this:
Added support for sd() for aggregate and window functions (#2887) (@edgararuiz)
Once ready, let me know, and I'll be glad to try out your fork before your submission.
Sorry for not doing anything on this one in the last week, I had use the chance to discuss it a user!2017 with Jim and Kirill. But I see you have worked hard in the meantime. Will check if something missing after sleeping over the Belgian Beer.
@edgararuiz:
Unit tests look easy enough to create. I suppose I should use the test connection cn <- simulate_odbc(type = "ACCESS")? It seems to do what I want when I test translate_sql(as.numeric(x), con = cn) (meaning it uses my translation).
Should I submit two pull requests? First for names_to_as() and then for the Access translation? names_to_as() will affect more than just Access so it might make sense that way.
Good question, you'll need to add simulate_odbc_access() to the simlulate.r script and then call it from your new test script
I'd suggest in PR because we can't really merge the translation w/o the names_to_as() change
If you fully qualify the table name, you can use the "as". This would be more generalizable.
SELECT [pat].[patient] as [patient] from pat
@dmenne, I like the thought here because I'd rather fully qualify names like this, it seems to be better practice and more robust. Here are a few points I thought of that might make this challenging:
1) We might should use `instead of []. I'm not sure how common it is to use brackets in other db's, and
SELECT \pat`.`patient` as `patient` from pat
still works great with Access!
2) Remember that this change would likely be used in all other databases as well. Do all databases support fully qualifying table names with a "." between the db name and the table name? (I'm assuming yes)
3) The change would likely take place in a different function than previously suggested. The names_to_as() function does not know about the db name, so its not as simple as before when i suggested dropping the "as" if the value of the ident object is the same as its name. Perhaps @edgararuiz has an idea of where we would make the change for this. Maybe the ident object?
4) While I like the verbose-ness of fully qualifying names, is it really necessary in the queries that we write with dplyr? I guess it could be needed when joining two tables together if they had two columns with the same name? Do you see any other need for it? If it looks like the change is too difficult, we should consider if it is even necessary over just dropping the "as".
@DavisVaughan
I just stumbled over the [] solution, and I have not looked well enough into the consequences. I thought that this was going to be a special Access driver anyway, but looks like parts from other solutions are kept. You decide....
@dmenne / @DavisVaughan - while researching the name_to_as() solution, I stumbled into the [] as an option, but I'm not sure how we could do this in indent because the the beginning and end characters are different, so we would have to come up with a modification to the ident() or maybe even a lower level function to get it to work. Can we start with stay on course and run a battery of test over the proposed name_to_as function? I have a repo that does some automated testing that we can leverage: https://github.com/rstudio/dbtest
I also found the sql_table_prefix() function that already exists. It's used in fully qualifying names when doing joins. It kind of does what we want.
library(dbplyr)
library(DBI)
cn <- dbConnect(odbc::odbc(), dsn = "access-odbc")
dbplyr:::sql_table_prefix(cn, var = ident("col1"), table = ident("table1"))
#> <SQL> `table1`.`col1`
I tried adding this to the sql_select.ACCESS function, when building out$select, by changing:
escape(select, collapse = ", ", con = con)
to:
escape(sql_table_prefix(con, select, from) ,
collapse = ", ",
con = con)
This simple implementation works with basic select queries, and has the benefit of being part of the ACCESS only implementation (doesn't affect the other db's!), but it fails with things like * because it tries to do `table1`.`*` and placing back ticks around the * is not something that works in access. Instead it wants `table1`.* without backticks. I also don't know how it would work with joins that automatically try and add (I think) "TABLE_LEFT" and "TABLE_RIGHT" as aliased table names (havent tried any examples).
_All that to say that it could be a solution, but I agree that we should try the initial names_to_as() change and run the large amount of tests over it._
I'll get it to you as soon as I can!
@edgararuiz
- Tests have been added
- NEWS has been updated
I think its ready for you to run through the gauntlet of tests and try out.
https://github.com/DavisVaughan/dbplyr
There are a few more issues I'd like to discuss regarding:
*_join- when joining two tables that have a column name in common (say, mtcars with itself), dbplyr will try and alias as `TBL_LEFT`.`cyl` AS `cyl.x` and `TBL_RIGHT`.`cyl` AS `cyl.y`. Access does not allow periods in field names :( see herefull_join. Uses coalesce. See issue below.- the
coalescefunction. Doesn't exist in access. Use NZ() instead. copy_touses TEMPORARY tables which doesn't exist in AccessCBool(akaas.logical) returns -1 for TRUE and 0 for FALSE)
I'll post on these later today hopefully.
Problem
left_join doesn't work right out of the box when joining two tables with columns of the same name (meaning if table1 has a column disp and table2 has a column disp). The reason is that suffixes are defaulted to c(".x", ".y") and Access can't use "." in column names.
This may or may not be a problem for other databases as well. I am not sure how many allow you to use a period in a column name.
Reprex
library(DBI)
library(dbplyr)
suppressPackageStartupMessages(library(dplyr))
cn <- dbConnect(odbc::odbc(), dsn = "dbplyr-testing")
cars <- tbl(cn, "mtcars")
cars2 <- tbl(cn, "mtcars2")
# Just mtcars
head(cars)
#> # Source: lazy query [?? x 11]
#> # Database: ACCESS
#> # 14.00.0000[admin@ACCESS/C:\Users\Administrator\Desktop\R\dbplyr-testing\dplyr-test-db.accdb]
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <int> <dbl> <int> <dbl> <dbl> <dbl> <int> <int> <int> <int>
#> 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
#> 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
#> 4 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
#> 5 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
#> 6 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
# A smaller version of mtcars
head(cars2)
#> # Source: lazy query [?? x 11]
#> # Database: ACCESS
#> # 14.00.0000[admin@ACCESS/C:\Users\Administrator\Desktop\R\dbplyr-testing\dplyr-test-db.accdb]
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <int> <dbl> <int> <dbl> <dbl> <dbl> <int> <int> <int> <int>
#> 1 14.3 8 360.0 245 3.21 3.57 15.84 0 0 3 4
#> 2 17.8 6 167.6 123 3.92 3.44 18.90 1 0 4 4
#> 3 16.4 8 275.8 180 3.07 4.07 17.40 0 0 3 3
#> 4 17.3 8 275.8 180 3.07 3.73 17.60 0 0 3 3
#> 5 15.2 8 275.8 180 3.07 3.78 18.00 0 0 3 3
#> 6 10.4 8 472.0 205 2.93 5.25 17.98 0 0 3 4
# Left join doesn't work because of the c(".x", ".y") suffix
left_join(cars, cars2, by = c("mpg", "cyl"))
#> Error: <SQL> 'SELECT TOP 10 *
#> FROM (SELECT `TBL_LEFT`.`mpg` AS `mpg`, `TBL_LEFT`.`cyl` AS `cyl`, `TBL_LEFT`.`disp` AS `disp.x`, `TBL_LEFT`.`hp` AS `hp.x`, `TBL_LEFT`.`drat` AS `drat.x`, `TBL_LEFT`.`wt` AS `wt.x`, `TBL_LEFT`.`qsec` AS `qsec.x`, `TBL_LEFT`.`vs` AS `vs.x`, `TBL_LEFT`.`am` AS `am.x`, `TBL_LEFT`.`gear` AS `gear.x`, `TBL_LEFT`.`carb` AS `carb.x`, `TBL_RIGHT`.`disp` AS `disp.y`, `TBL_RIGHT`.`hp` AS `hp.y`, `TBL_RIGHT`.`drat` AS `drat.y`, `TBL_RIGHT`.`wt` AS `wt.y`, `TBL_RIGHT`.`qsec` AS `qsec.y`, `TBL_RIGHT`.`vs` AS `vs.y`, `TBL_RIGHT`.`am` AS `am.y`, `TBL_RIGHT`.`gear` AS `gear.y`, `TBL_RIGHT`.`carb` AS `carb.y`
#> FROM `mtcars` AS `TBL_LEFT`
#> LEFT JOIN `mtcars2` AS `TBL_RIGHT`
#> ON (`TBL_LEFT`.`mpg` = `TBL_RIGHT`.`mpg` AND `TBL_LEFT`.`cyl` = `TBL_RIGHT`.`cyl`)
#> ) `owhsmsymuo`'
#> nanodbc/nanodbc.cpp:1587: 42000: [Microsoft][ODBC Microsoft Access Driver] 'disp.x' is not a valid name. Make sure that it does not include invalid characters or punctuation and that it is not too long.
# It works if you force a suffix of c("_x", "_y")
left_join(cars, cars2, by = c("mpg", "cyl"), suffix = c("_x", "_y"))
#> # Source: lazy query [?? x 20]
#> # Database: ACCESS
#> # 14.00.0000[admin@ACCESS/C:\Users\Administrator\Desktop\R\dbplyr-testing\dplyr-test-db.accdb]
#> mpg cyl disp_x hp_x drat_x wt_x qsec_x vs_x am_x gear_x carb_x
#> <dbl> <int> <dbl> <int> <dbl> <dbl> <dbl> <int> <int> <int> <int>
#> 1 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
#> 2 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
#> 3 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> 4 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
#> 5 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
#> 6 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
#> 7 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
#> 8 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> 9 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
#> 10 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
#> # ... with more rows, and 9 more variables: disp_y <dbl>, hp_y <int>,
#> # drat_y <dbl>, wt_y <dbl>, qsec_y <dbl>, vs_y <int>, am_y <int>,
#> # gear_y <int>, carb_y <int>
Potential solutions
1) Since left_join is a generic, create a new left_join.tbl_dbi() (currently left_join.tbl_lazy is used) that uses c("_x", "_y"). _This would affect all databases._
2) Just tell Access users to specify the argument suffix = c("_x", "_y") in this specific case of a left_join() where you have column names from each table that are the same.
The choice is up to you, but I think it kind of depends on if this is a problem in other databases as well.
As an argument _for_ the change to c("_x", "_y"), it seems to be bad practice to add "." in field names or database names. See here
Edited: Update
This does not seem to be a problem with MySQL.
library(DBI)
suppressPackageStartupMessages(library(dplyr))
library(dbplyr)
#>
#> Attaching package: 'dbplyr'
#> The following objects are masked from 'package:dplyr':
#>
#> ident, sql
cn <- dbConnect(drv = RMySQL::MySQL(),
username = "user1",
password = "testpassword",
host = "davisdbinstance.crarljboc8to.us-west-2.rds.amazonaws.com",
port = 3306,
dbname = "firstdb")
apple <- tbl(cn, "apple")
ibm <- tbl(cn, "ibm")
head(apple, 2)
#> # Source: lazy query [?? x 8]
#> # Database: mysql 5.6.27-log
#> # [[email protected]:/firstdb]
#> row_names date open high low close volume adjusted
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 2007-01-03 86.29 86.58 81.90 83.80 309579900 10.90416
#> 2 2 2007-01-04 84.05 85.95 83.82 85.66 211815100 11.14619
head(ibm, 2)
#> # Source: lazy query [?? x 7]
#> # Database: mysql 5.6.27-log
#> # [[email protected]:/firstdb]
#> date open high low close volume adjusted
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2007-01-03 97.18 98.40 96.26 97.27 9196800 78.35465
#> 2 2007-01-04 97.25 98.79 96.88 98.31 10524500 79.19241
left_join(apple, ibm, by = "date")
#> # Source: lazy query [?? x 14]
#> # Database: mysql 5.6.27-log
#> # [[email protected]:/firstdb]
#> row_names date open.x high.x low.x close.x volume.x adjusted.x
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 2007-01-03 86.29 86.58 81.90 83.80 309579900 10.90416
#> 2 2 2007-01-04 84.05 85.95 83.82 85.66 211815100 11.14619
#> 3 3 2007-01-05 85.77 86.20 84.40 85.05 208685400 11.06681
#> 4 4 2007-01-08 85.96 86.53 85.28 85.47 199276700 11.12147
#> 5 5 2007-01-09 86.45 92.98 85.15 92.57 837324600 12.04533
#> 6 6 2007-01-10 94.75 97.80 93.45 97.00 738220000 12.62176
#> 7 7 2007-01-11 95.94 96.78 95.10 95.80 360063200 12.46562
#> 8 8 2007-01-12 94.59 95.06 93.23 94.62 328172600 12.31207
#> 9 9 2007-01-16 95.68 97.25 95.45 97.10 311019100 12.63477
#> 10 10 2007-01-17 97.56 97.60 94.82 94.95 411565000 12.35501
#> # ... with more rows, and 6 more variables: open.y <dbl>, high.y <dbl>,
#> # low.y <dbl>, close.y <dbl>, volume.y <dbl>, adjusted.y <dbl>
@edgararuiz, I've made some progress here. Your dbtest package wouldn't work for me out of the box (because of Access), but I made a few changes and got things running. Looks like most of the tests pass, with a few exceptions that I looked into, and document below.
........EE.......EW........................FF.......EEEEEEEEEEEEEEEEE
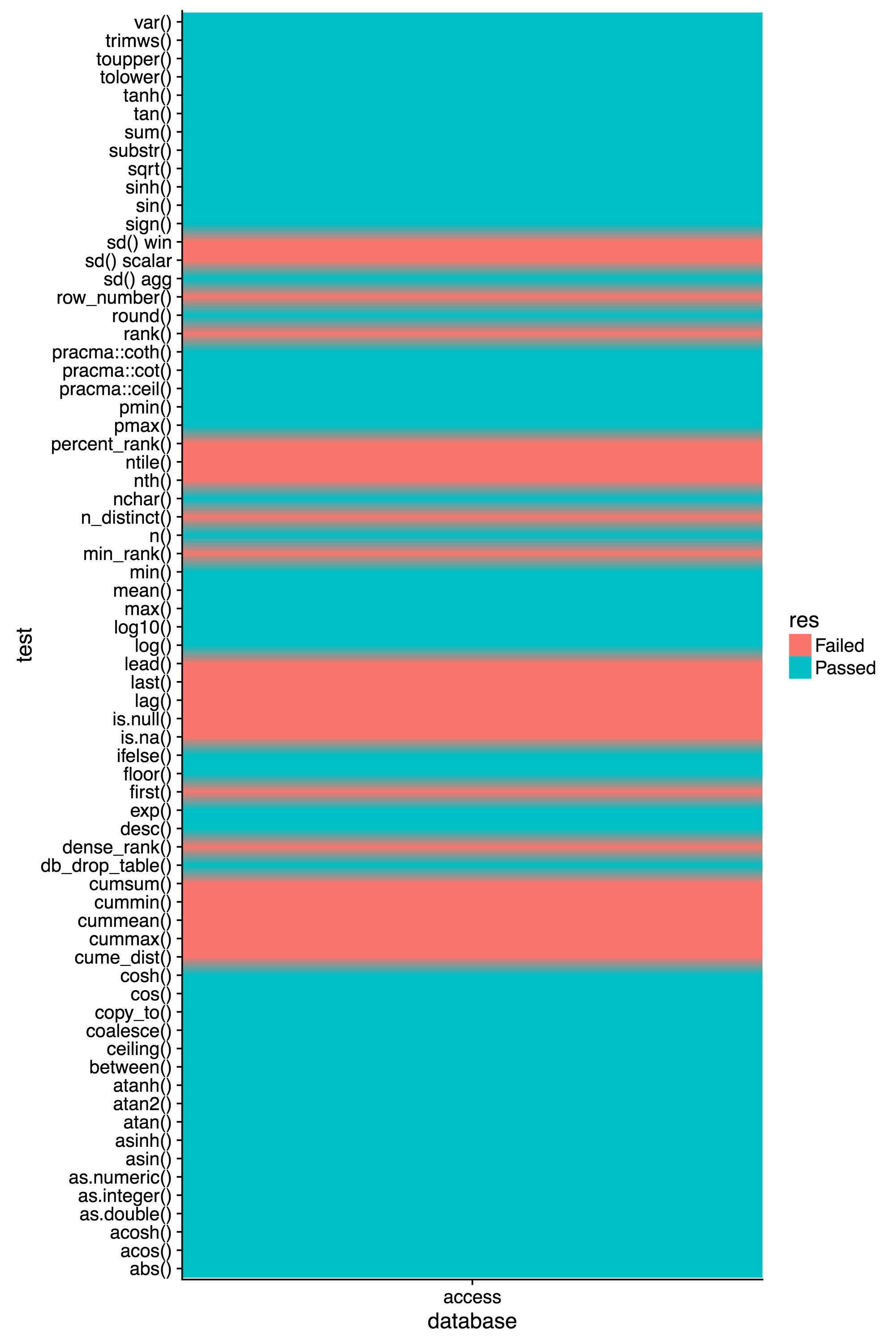
Failing tests
1) n_distinct() is not easily supported in Access. Most implementations do Select Count(Distinct *) ..., but unfortunately Access can't do that. I used sql_not_supported() to let the user know. Answers online say to use Select Count(*) From (Select Distinct * From table_name) as T, but that requires knowing the table name, which you don't know inside the sql_translator() function.
2) In test-3-scalar.R, when running the test sd() scalar, I believe it actually gets passed through as a window function, not a scalar. Access does not support window functions, and I have used win_absent(sd) in the win part of sql_translator(), and I get the error, "Window function sd() is not supported by this database" when I try to run that test, which leads me to believe it is being seen as a window function.
3) There is an issue with my implementation of atanh() in that if you pass in 1 or -1 you will get a division by 0 error. In R it returns Inf or -Inf, but I can't find a way to make Access do this.
4) is.na() and is.null() should both return logicals, but I'm not sure how to do this. The Boolean data type in Access is just a column that holds -1 for TRUE and 0 for FALSE. I'm not sure how to get R to understand that that is a logical. Right now, I do this IIF(ISNULL(x), 1, 0) so instead of using Access's odd boolean data type, I just put in an integer 1 or 0 if it is null/na.
5) The rest of the errors that you are seeing (all of the ones at the end of the minimal output) are all window functions. As far as I know, Access does not support them, so all of them have been noted using win_absent().
Full test code
# Packages
library(DBI)
library(dbplyr)
library(dplyr, warn.conflicts = FALSE)
library(dbtest, warn.conflicts = FALSE)
#> Warning: replacing previous import 'dplyr::contains' by 'purrr::contains'
#> when loading 'dbtest'
#> Warning: replacing previous import 'dplyr::order_by' by 'purrr::order_by'
#> when loading 'dbtest'
#> Warning: replacing previous import 'purrr::is_null' by 'testthat::is_null'
#> when loading 'dbtest'
#> Warning: replacing previous import 'dplyr::matches' by 'testthat::matches'
#> when loading 'dbtest'
library(pracma)
# wd has to be set for test_database config to find the config.yml
setwd("C:/Users/Administrator/Desktop/R/dbtest")
# Three of the db_* functions have to have Access specific implementations
# db_write_table.ACCESS - odbc package does not support writing to Access yet. Open issue on that package
# db_copy_to.ACCESS - same as above
# db_drop_table.ACCESS - "IF EXISTS" is not an Access SQL command, so this had to be rewritten too
source("../db-write-table.R")
# Now it works.
# There are a few additional changes that have been made to your dbtest package so that I could even run this function
x <- test_database(databases = "access")
#>
#> Attaching package: 'testthat'
#> The following object is masked from 'package:dplyr':
#>
#> matches
#> ........EE.......EW........................FF.......EEEEEEEEEEEEEEEEE.
# This is ugly, but its all there
x
#> file context test nb failed skipped error
#> 1 test-1-start.R initialize copy_to() 1 0 FALSE FALSE
#> 2 test-2-agg.R base_agg n() 1 0 FALSE FALSE
#> 3 test-2-agg.R base_agg mean() 1 0 FALSE FALSE
#> 4 test-2-agg.R base_agg var() 1 0 FALSE FALSE
#> 5 test-2-agg.R base_agg sd() agg 1 0 FALSE FALSE
#> 6 test-2-agg.R base_agg sum() 1 0 FALSE FALSE
#> 7 test-2-agg.R base_agg min() 1 0 FALSE FALSE
#> 8 test-2-agg.R base_agg max() 1 0 FALSE FALSE
#> 9 test-2-agg.R base_agg n_distinct() 0 0 FALSE TRUE
#> 10 test-3-scalar.R base_scalar sd() scalar 0 0 FALSE TRUE
#> 11 test-3-scalar.R base_scalar abs() 1 0 FALSE FALSE
#> 12 test-3-scalar.R base_scalar acos() 1 0 FALSE FALSE
#> 13 test-3-scalar.R base_scalar acosh() 1 0 FALSE FALSE
#> 14 test-3-scalar.R base_scalar asin() 1 0 FALSE FALSE
#> 15 test-3-scalar.R base_scalar asinh() 1 0 FALSE FALSE
#> 16 test-3-scalar.R base_scalar atan() 1 0 FALSE FALSE
#> 17 test-3-scalar.R base_scalar atan2() 1 0 FALSE FALSE
#> 18 test-3-scalar.R base_scalar atanh() 2 0 FALSE FALSE
#> 19 test-3-scalar.R base_scalar pracma::ceil() 1 0 FALSE FALSE
#> 20 test-3-scalar.R base_scalar ceiling() 1 0 FALSE FALSE
#> 21 test-3-scalar.R base_scalar cos() 1 0 FALSE FALSE
#> 22 test-3-scalar.R base_scalar cosh() 1 0 FALSE FALSE
#> 23 test-3-scalar.R base_scalar pracma::cot() 1 0 FALSE FALSE
#> 24 test-3-scalar.R base_scalar pracma::coth() 1 0 FALSE FALSE
#> 25 test-3-scalar.R base_scalar exp() 1 0 FALSE FALSE
#> 26 test-3-scalar.R base_scalar floor() 1 0 FALSE FALSE
#> 27 test-3-scalar.R base_scalar log() 1 0 FALSE FALSE
#> 28 test-3-scalar.R base_scalar log10() 1 0 FALSE FALSE
#> 29 test-3-scalar.R base_scalar round() 1 0 FALSE FALSE
#> 30 test-3-scalar.R base_scalar sign() 1 0 FALSE FALSE
#> 31 test-3-scalar.R base_scalar sin() 1 0 FALSE FALSE
#> 32 test-3-scalar.R base_scalar sinh() 1 0 FALSE FALSE
#> 33 test-3-scalar.R base_scalar sqrt() 1 0 FALSE FALSE
#> 34 test-3-scalar.R base_scalar tan() 1 0 FALSE FALSE
#> 35 test-3-scalar.R base_scalar tanh() 1 0 FALSE FALSE
#> 36 test-3-scalar.R base_scalar tolower() 1 0 FALSE FALSE
#> 37 test-3-scalar.R base_scalar toupper() 1 0 FALSE FALSE
#> 38 test-3-scalar.R base_scalar trimws() 1 0 FALSE FALSE
#> 39 test-3-scalar.R base_scalar nchar() 1 0 FALSE FALSE
#> 40 test-3-scalar.R base_scalar substr() 1 0 FALSE FALSE
#> 41 test-3-scalar.R base_scalar ifelse() 1 0 FALSE FALSE
#> 42 test-3-scalar.R base_scalar desc() 1 0 FALSE FALSE
#> 43 test-3-scalar.R base_scalar is.null() 1 1 FALSE FALSE
#> 44 test-3-scalar.R base_scalar is.na() 1 1 FALSE FALSE
#> 45 test-3-scalar.R base_scalar coalesce() 1 0 FALSE FALSE
#> 46 test-3-scalar.R base_scalar as.numeric() 1 0 FALSE FALSE
#> 47 test-3-scalar.R base_scalar as.double() 1 0 FALSE FALSE
#> 48 test-3-scalar.R base_scalar as.integer() 1 0 FALSE FALSE
#> 49 test-3-scalar.R base_scalar between() 1 0 FALSE FALSE
#> 50 test-3-scalar.R base_scalar pmin() 1 0 FALSE FALSE
#> 51 test-3-scalar.R base_scalar pmax() 1 0 FALSE FALSE
#> 52 test-4-win.R base_win row_number() 0 0 FALSE TRUE
#> 53 test-4-win.R base_win min_rank() 0 0 FALSE TRUE
#> 54 test-4-win.R base_win rank() 0 0 FALSE TRUE
#> 55 test-4-win.R base_win dense_rank() 0 0 FALSE TRUE
#> 56 test-4-win.R base_win percent_rank() 0 0 FALSE TRUE
#> 57 test-4-win.R base_win cume_dist() 0 0 FALSE TRUE
#> 58 test-4-win.R base_win ntile() 0 0 FALSE TRUE
#> 59 test-4-win.R base_win first() 0 0 FALSE TRUE
#> 60 test-4-win.R base_win last() 0 0 FALSE TRUE
#> 61 test-4-win.R base_win nth() 0 0 FALSE TRUE
#> 62 test-4-win.R base_win lead() 0 0 FALSE TRUE
#> 63 test-4-win.R base_win lag() 0 0 FALSE TRUE
#> 64 test-4-win.R base_win cummean() 0 0 FALSE TRUE
#> 65 test-4-win.R base_win cumsum() 0 0 FALSE TRUE
#> 66 test-4-win.R base_win cummin() 0 0 FALSE TRUE
#> 67 test-4-win.R base_win sd() win 0 0 FALSE TRUE
#> 68 test-4-win.R base_win cummax() 0 0 FALSE TRUE
#> 69 test-5-end.R cleanup db_drop_table() 1 0 FALSE FALSE
#> warning user system real database
#> 1 0 0.06 0.02 0.08 access
#> 2 0 0.21 0.00 0.20 access
#> 3 0 0.18 0.00 0.19 access
#> 4 0 0.05 0.00 0.04 access
#> 5 0 0.04 0.00 0.05 access
#> 6 0 0.05 0.00 0.05 access
#> 7 0 0.03 0.00 0.03 access
#> 8 0 0.05 0.00 0.05 access
#> 9 0 0.03 0.00 0.03 access
#> 10 0 0.05 0.00 0.04 access
#> 11 0 0.09 0.00 0.11 access
#> 12 0 0.05 0.00 0.05 access
#> 13 0 0.06 0.00 0.06 access
#> 14 0 0.05 0.00 0.05 access
#> 15 0 0.04 0.00 0.05 access
#> 16 0 0.05 0.00 0.06 access
#> 17 0 0.05 0.00 0.05 access
#> 18 1 0.04 0.00 0.04 access
#> 19 0 0.05 0.00 0.05 access
#> 20 0 0.05 0.00 0.05 access
#> 21 0 0.05 0.00 0.04 access
#> 22 0 0.04 0.00 0.05 access
#> 23 0 0.05 0.00 0.05 access
#> 24 0 0.06 0.00 0.06 access
#> 25 0 0.05 0.00 0.05 access
#> 26 0 0.05 0.00 0.04 access
#> 27 0 0.04 0.00 0.05 access
#> 28 0 0.06 0.00 0.06 access
#> 29 0 0.05 0.00 0.05 access
#> 30 0 0.05 0.00 0.05 access
#> 31 0 0.05 0.00 0.04 access
#> 32 0 0.04 0.00 0.05 access
#> 33 0 0.05 0.00 0.05 access
#> 34 0 0.05 0.00 0.04 access
#> 35 0 0.06 0.00 0.07 access
#> 36 0 0.05 0.00 0.04 access
#> 37 0 0.04 0.00 0.05 access
#> 38 0 0.05 0.00 0.05 access
#> 39 0 0.05 0.00 0.04 access
#> 40 0 0.04 0.00 0.05 access
#> 41 0 0.07 0.00 0.06 access
#> 42 0 0.04 0.00 0.05 access
#> 43 0 0.05 0.00 0.05 access
#> 44 0 0.05 0.00 0.04 access
#> 45 0 0.04 0.00 0.05 access
#> 46 0 0.05 0.00 0.05 access
#> 47 0 0.05 0.00 0.04 access
#> 48 0 0.04 0.00 0.05 access
#> 49 0 0.05 0.00 0.05 access
#> 50 0 0.04 0.00 0.05 access
#> 51 0 0.03 0.00 0.05 access
#> 52 0 0.02 0.00 0.02 access
#> 53 0 0.03 0.00 0.03 access
#> 54 0 0.01 0.00 0.02 access
#> 55 0 0.03 0.00 0.03 access
#> 56 0 0.03 0.00 0.03 access
#> 57 0 0.02 0.00 0.01 access
#> 58 0 0.03 0.00 0.04 access
#> 59 0 0.10 0.00 0.10 access
#> 60 0 0.01 0.00 0.02 access
#> 61 0 0.02 0.00 0.02 access
#> 62 0 0.03 0.00 0.03 access
#> 63 0 0.03 0.00 0.03 access
#> 64 0 0.01 0.00 0.01 access
#> 65 0 0.02 0.00 0.02 access
#> 66 0 0.02 0.00 0.03 access
#> 67 0 0.01 0.00 0.02 access
#> 68 0 0.03 0.00 0.03 access
#> 69 0 0.00 0.00 0.01 access
#> result
#> 1 {\n ...\n} produced .\n
#> 2 manip(db) not equal to manip(local).\nEqual\n
#> 3 manip(db) not equal to manip(local).\nEqual\n
#> 4 manip(db) not equal to manip(local).\nEqual\n
#> 5 manip(db) not equal to manip(local).\nEqual\n
#> 6 manip(db) not equal to manip(local).\nEqual\n
#> 7 manip(db) not equal to manip(local).\nEqual\n
#> 8 manip(db) not equal to manip(local).\nEqual\n
#> 9 N_DISTINCT is not available in this SQL variant
#> 10 Window function `sd()` is not supported by this database
#> 11 manip(db) not equal to manip(local).\nEqual\n
#> 12 manip(db) not equal to manip(local).\nEqual\n
#> 13 manip(db) not equal to manip(local).\nEqual\n
#> 14 manip(db) not equal to manip(local).\nEqual\n
#> 15 manip(db) not equal to manip(local).\nEqual\n
#> 16 manip(db) not equal to manip(local).\nEqual\n
#> 17 manip(db) not equal to manip(local).\nEqual\n
#> 18 nanodbc/nanodbc.cpp:2525: 22012: [Microsoft][ODBC Microsoft Access Driver]Division by zero
#> 19 manip(db) not equal to manip(local).\nEqual\n
#> 20 manip(db) not equal to manip(local).\nEqual\n
#> 21 manip(db) not equal to manip(local).\nEqual\n
#> 22 manip(db) not equal to manip(local).\nEqual\n
#> 23 manip(db) not equal to manip(local).\nEqual\n
#> 24 manip(db) not equal to manip(local).\nEqual\n
#> 25 manip(db) not equal to manip(local).\nEqual\n
#> 26 manip(db) not equal to manip(local).\nEqual\n
#> 27 manip(db) not equal to manip(local).\nEqual\n
#> 28 manip(db) not equal to manip(local).\nEqual\n
#> 29 manip(db) not equal to manip(local).\nEqual\n
#> 30 manip(db) not equal to manip(local).\nEqual\n
#> 31 manip(db) not equal to manip(local).\nEqual\n
#> 32 manip(db) not equal to manip(local).\nEqual\n
#> 33 manip(db) not equal to manip(local).\nEqual\n
#> 34 manip(db) not equal to manip(local).\nEqual\n
#> 35 manip(db) not equal to manip(local).\nEqual\n
#> 36 manip(db) not equal to manip(local).\nEqual\n
#> 37 manip(db) not equal to manip(local).\nEqual\n
#> 38 manip(db) not equal to manip(local).\nEqual\n
#> 39 manip(db) not equal to manip(local).\nEqual\n
#> 40 manip(db) not equal to manip(local).\nEqual\n
#> 41 manip(db) not equal to manip(local).\nEqual\n
#> 42 manip(db) not equal to manip(local).\nEqual\n
#> 43 manip(db) not equal to manip(local).\nTypes not compatible: integer vs logical\n
#> 44 manip(db) not equal to manip(local).\nTypes not compatible: integer vs logical\n
#> 45 manip(db) not equal to manip(local).\nEqual\n
#> 46 manip(db) not equal to manip(local).\nEqual\n
#> 47 manip(db) not equal to manip(local).\nEqual\n
#> 48 manip(db) not equal to manip(local).\nEqual\n
#> 49 manip(db) not equal to manip(local).\nEqual\n
#> 50 manip(db) not equal to manip(local).\nEqual\n
#> 51 manip(db) not equal to manip(local).\nEqual\n
#> 52 Window function `row_number()` is not supported by this database
#> 53 Window function `min_rank()` is not supported by this database
#> 54 Window function `rank()` is not supported by this database
#> 55 Window function `dense_rank()` is not supported by this database
#> 56 Window function `percent_rank()` is not supported by this database
#> 57 Window function `cume_dist()` is not supported by this database
#> 58 Window function `ntile()` is not supported by this database
#> 59 Window function `first()` is not supported by this database
#> 60 Window function `last()` is not supported by this database
#> 61 Window function `nth()` is not supported by this database
#> 62 Window function `lead()` is not supported by this database
#> 63 Window function `lag()` is not supported by this database
#> 64 Window function `cummean()` is not supported by this database
#> 65 Window function `cumsum()` is not supported by this database
#> 66 Window function `cummin()` is not supported by this database
#> 67 Window function `sd()` is not supported by this database
#> 68 Window function `cummax()` is not supported by this database
#> 69 {\n ...\n} produced .\n
#> call
#> 1 expect_silent({\n dbplyr::db_copy_to(con = con, table = table_name, values = test_table, temporary = FALSE, types = c(fld_factor = "VARCHAR", fld_datetime = "DATETIME", fld_date = "DATETIME", fld_time = "VARCHAR", fld_binary = "LONG", fld_integer = "LONG", fld_double = "DOUBLE", fld_character = "VARCHAR", fld_logical = "LONG"))\n})
#> 2 expect_summarise_equivalent(n())
#> 3 expect_summarise_equivalent(mean(fld_double))
#> 4 expect_summarise_equivalent(var(fld_double))
#> 5 expect_summarise_equivalent(sd(fld_double))
#> 6 expect_summarise_equivalent(sum(fld_double))
#> 7 expect_summarise_equivalent(min(fld_double))
#> 8 expect_summarise_equivalent(max(fld_double))
#> 9 expect_summarise_equivalent(n_distinct(fld_double))
#> 10 expect_mutate_equivalent(sd(fld_double))
#> 11 expect_mutate_equivalent(abs(fld_double))
#> 12 expect_mutate_equivalent(acos(fld_binary))
#> 13 expect_mutate_equivalent(acosh(fld_double))
#> 14 expect_mutate_equivalent(asin(fld_binary))
#> 15 expect_mutate_equivalent(asinh(fld_double))
#> 16 expect_mutate_equivalent(atan(fld_double))
#> 17 expect_mutate_equivalent(atan2(fld_double, 1))
#> 18 expect_mutate_equivalent(atanh(fld_binary))
#> 19 expect_mutate_equivalent(ceil(fld_double))
#> 20 expect_mutate_equivalent(ceiling(fld_double))
#> 21 expect_mutate_equivalent(cos(fld_binary))
#> 22 expect_mutate_equivalent(cosh(fld_binary))
#> 23 expect_mutate_equivalent(cot(fld_double))
#> 24 expect_mutate_equivalent(coth(fld_double))
#> 25 expect_mutate_equivalent(exp(fld_double))
#> 26 expect_mutate_equivalent(floor(fld_double))
#> 27 expect_mutate_equivalent(log(fld_double))
#> 28 expect_mutate_equivalent(log10(fld_double))
#> 29 expect_mutate_equivalent(round(fld_double))
#> 30 expect_mutate_equivalent(sign(fld_double))
#> 31 expect_mutate_equivalent(sin(fld_double))
#> 32 expect_mutate_equivalent(sinh(fld_double))
#> 33 expect_mutate_equivalent(sqrt(fld_double))
#> 34 expect_mutate_equivalent(tan(fld_double))
#> 35 expect_mutate_equivalent(tanh(fld_double))
#> 36 expect_mutate_equivalent(tolower(fld_character))
#> 37 expect_mutate_equivalent(toupper(fld_character))
#> 38 expect_mutate_equivalent(trimws(fld_character))
#> 39 expect_mutate_equivalent(nchar(fld_character))
#> 40 expect_mutate_equivalent(substr(fld_character, 1, 1))
#> 41 expect_mutate_equivalent(ifelse(fld_binary == 1, "Yes", "No"))
#> 42 expect_mutate_equivalent(fld_character)
#> 43 expect_mutate_equivalent(is.null(fld_character))
#> 44 expect_mutate_equivalent(is.na(fld_character))
#> 45 expect_mutate_equivalent(coalesce(fld_character, fld_character))
#> 46 expect_mutate_equivalent(as.numeric(fld_logical))
#> 47 expect_mutate_equivalent(as.double(fld_logical))
#> 48 expect_mutate_equivalent(as.integer(fld_double))
#> 49 expect_mutate_equivalent(fld_double)
#> 50 expect_mutate_equivalent(pmin(fld_binary, fld_integer))
#> 51 expect_mutate_equivalent(pmax(fld_binary, fld_integer))
#> 52 expect_window_equivalent(row_number(fld_double))
#> 53 expect_window_equivalent(min_rank(fld_double))
#> 54 expect_window_equivalent(rank(fld_double))
#> 55 expect_window_equivalent(dense_rank(fld_double))
#> 56 expect_window_equivalent(percent_rank(fld_double))
#> 57 expect_window_equivalent(cume_dist(fld_double))
#> 58 expect_window_equivalent(ntile(fld_double, 2))
#> 59 expect_window_equivalent(first(fld_integer))
#> 60 expect_window_equivalent(last(fld_integer))
#> 61 expect_window_equivalent(nth(fld_integer, 5))
#> 62 expect_window_equivalent(lead(fld_integer))
#> 63 expect_window_equivalent(lag(fld_integer))
#> 64 expect_window_equivalent(cummean(fld_double))
#> 65 expect_window_equivalent(cumsum(fld_double))
#> 66 expect_window_equivalent(cummin(fld_double))
#> 67 expect_window_equivalent(sd(fld_double))
#> 68 expect_window_equivalent(cummax(fld_double))
#> 69 expect_silent({\n dplyr::db_drop_table(con, table_name, force = TRUE)\n})
#> res
#> 1 Passed
#> 2 Passed
#> 3 Passed
#> 4 Passed
#> 5 Passed
#> 6 Passed
#> 7 Passed
#> 8 Passed
#> 9 Failed
#> 10 Failed
#> 11 Passed
#> 12 Passed
#> 13 Passed
#> 14 Passed
#> 15 Passed
#> 16 Passed
#> 17 Passed
#> 18 Passed
#> 19 Passed
#> 20 Passed
#> 21 Passed
#> 22 Passed
#> 23 Passed
#> 24 Passed
#> 25 Passed
#> 26 Passed
#> 27 Passed
#> 28 Passed
#> 29 Passed
#> 30 Passed
#> 31 Passed
#> 32 Passed
#> 33 Passed
#> 34 Passed
#> 35 Passed
#> 36 Passed
#> 37 Passed
#> 38 Passed
#> 39 Passed
#> 40 Passed
#> 41 Passed
#> 42 Passed
#> 43 Failed
#> 44 Failed
#> 45 Passed
#> 46 Passed
#> 47 Passed
#> 48 Passed
#> 49 Passed
#> 50 Passed
#> 51 Passed
#> 52 Failed
#> 53 Failed
#> 54 Failed
#> 55 Failed
#> 56 Failed
#> 57 Failed
#> 58 Failed
#> 59 Failed
#> 60 Failed
#> 61 Failed
#> 62 Failed
#> 63 Failed
#> 64 Failed
#> 65 Failed
#> 66 Failed
#> 67 Failed
#> 68 Failed
#> 69 Passed
Hi @DavisVaughan ,
Sorry for taking some time in getting back with you. I have some more work to do on dbtest, but I think that the results you are posting are acceptable. The points you raised in the Failing Tests are in line with other translations I've worked on, specially number 4.
Is there any outstanding work you believe is needed still?
Hey @edgararuiz, no problem.
Just added a few more translation tests and cleaned up the syntax a bit. I think it is ready to go if we are okay with the errors above. There are a few things you should know:
General
1) The branch is feature-access-sql-translation on my fork here. I can do a pull request whenever you want, but know that that branch itself is a branch off of the feature-select-names branch that is already an open pull request here. I was thinking that it would be accepted first, but I get that you guys are busy.
2) Access only supports sine, cosine, and arctan natively, so I had to get creative to construct the other trig functions from those 3. I included a link to someone who had done most of the work for me in the comments of that section. They look messy but they work and I couldn't think of a better way.
3) While working with the database, I discovered three db_* functions that don't work, and I rolled my own versions that do. I've created a Gist with that code here and we can talk about including it too if needed.
1. `db_drop_table` - Originally uses `IF EXISTS` if "forcing" the drop is required. Access doesn't support this. Rewrote to ask the user if they want to drop, and require them to specify `force = TRUE` if they do.
2. `db_copy_to` - As of now, temporary tables aren't supported in Access, so I had to turn that option off.
3. `db_write_table` - This is a big one. Already posted [here](https://github.com/rstats-db/odbc/issues/79), but `dbWriteTable()` does not yet work with Access `odbc` connections. Fortunately, I found Hadley's solution for [writing tables to MySQL](https://github.com/tidyverse/dbplyr/blob/master/R/db-mysql.r#L80) and Access can actually do a similar thing!
Other known errors
I thought I would bring these back up to include on the db.rstudio.com page
1) As mentioned in an earlier comment, when joining two tables with the same column names, you have to specify suffix = c("_x", "_y") over the default of suffix = c(".x", ".y") because there cannot be . in Access column names.
2) full_join uses the coalesce function in the SQL that it builds. This is not supported in Access. There is an alternative of using IFF(Is.Null(x), y, x) but the sql_coalesce() function in dbplyr is not generic and I don't think it ever sees the connection object. That alternative solution would also only work with a max of two tables being "coalesced" at once (probably fine with joins but maybe a limitation elsewhere). Below is the current sql_coalesce() function for reference:
sql_coalesce <- function(...) {
vars <- sql_vector(list(...), parens = FALSE, collapse = ", ")
build_sql("coalesce(", vars, ")")
}
@DavisVaughan - Unless you believe that there is some other feature you'd like to add, I think we are ready to go with a PR.
I tried it this morning, and could not get db_write_table to work (error message on request, currently not on my devel machine). Will the Gist mentioned above be included? I think this are heavy-used functions, and the equivalents work on RODBC.
@dmenne - were you able to try DBI::dbWriteTable() as well?
library(odbc)
library(dplyr)
library(dbplyr)
library(DBI)
cs = "Driver=Microsoft Access Driver (*.mdb, *.accdb);DBQ=./test.accdb"
con = dbConnect(odbc::odbc(), .connection_string = cs)
DBI::dbListTables(con)
# Ok
a = tbl(con, "pat")
a %>% show_query()
a
# Ok
translate_sql(first %like% "Had%", con)
src <- src_dbi(con)
src %>% tbl("pat")
# Add some data
DBI::dbWriteTable(con, "cars", mtcars)
Error in new_result(connection@ptr, statement) :
nanodbc/nanodbc.cpp:1344: 42000: [Microsoft][ODBC-Treiber für Microsoft Access] Syntaxfehler in CREATE TABLE-Anweisung.
Ok, my German is a little rusty 😄 , but it looks like an incorrect syntax error is being returned at the DBI level, which means that we'll need a custom solution for this. The three options I see are to create a customization in either odbc, DBI or dbplyr::db_create_table.
My suggestion would be not 3, because there will be others that will try to use DBI::dbWriteTable directly, so a fix should be done at a lower level than dbplyr. This means that the current translation should be sent over as a PR as-is, so folks can start using the 'read-only' capabilities, while we work on a solution for writing tables.
@hadley - Would you mind weighing in?
Another variant:
library(odbc)
library(dplyr)
library(dbplyr)
library(DBI)
cs = "Driver=Microsoft Access Driver (*.mdb, *.accdb);DBQ=./test.accdb"
con = dbConnect(odbc::odbc(), .connection_string = cs)
src = src_dbi(con)
pt = as.data.frame(src %>% tbl("pat"))
pt$ID = pt$ID + 100L # Make unique
# Add some data
DBI::dbWriteTable(con, "pat", pt, append = TRUE )
Error in result_insert_dataframe(rs@ptr, values) :
nanodbc/nanodbc.cpp:1791: HY104: [Microsoft][ODBC-Treiber für Microsoft Access]Ungültiger Genauigkeitswert.
"Invalid precision" for the germycapped.
Ok, that sounds like the pat and pt field definitions may be mismatched, either inside R or when the translation occurs. This reminds me that we will also need a Data Type translation PR sent over to the odbc package: https://github.com/rstats-db/odbc/blob/master/R/DataTypes.R
@dmenne - Can you look at the variable types returned by tbl(con, "pat") and pt? This should tell us if the differences are within R
I forgot to mention it, because it looked clean.
src = src_dbi(con)
pt = as.data.frame(src %>% tbl("pat"))
#'data.frame': 3 obs. of 2 variables:
# $ ID : int 1 2 3
# $ patient: chr "a" "b" "c"
tbl(con, "pat")
# Source: table<pat> [?? x 2]
# Database: ACCESS 12.00.0000[admin@ACCESS/./test.accdb]
# ID patient
# <int> <chr>
# 1 1 a
# 2 2 b
# 3 3 c
I'm curios what will happen if you map the right data types in your script using this function:
odbcDataType.default <- function(con, obj, ...) {
switch_type(obj,
factor = "VARCHAR(255)",
datetime = "TIMESTAMP",
date = "DATE",
time = "TIME",
binary = "VARBINARY(255)",
integer = "INTEGER",
double = "DOUBLE PRECISION",
character = "VARCHAR(255)",
logical = "BIT", # only valid if DB supports Null fields
list = "VARCHAR(255)",
stop("Unsupported type", call. = FALSE)
)
}
You need to change default to ACCESS, and change the right side of each data type argument to match Access' types
You may already know them by memory, but just in case, here's a list of Access datatypes I came accross: https://www.w3schools.com/sql/sql_datatypes.asp
Corrected, see below
Please check if I downloaded the correct version from github. Strongly simplfied the types-
library(odbc)
library(dplyr)
library(dbplyr)
library(DBI)
# to be safe:
#devtools::install_github("DavisVaughan/dbplyr@feature-access-sql-translation", force = TRUE)
cs = "Driver=Microsoft Access Driver (*.mdb, *.accdb);DBQ=./test.accdb"
con = dbConnect(odbc::odbc(), .connection_string = cs)
src = src_dbi(con)
pat = tbl(con, "pat")
pat$ID = pat$ID + 100L # Make unique (ID is primary)
odbcDataType.ACCESS <- function(con, obj, ...) {
switch_type(obj,
integer = "Integer",
character = "Text",
stop("Unsupported type", call. = FALSE)
)
}
# Add some data
DBI::dbWriteTable(con, "pat", pt, append = TRUE )
#other attached packages:
# [1] DBI_0.7 dbplyr_1.1.0.9000 dplyr_0.7.2 odbc_1.1.1.9000
<< Funny, it seems to listen to my orders to write English>>
Nevertheless, this looks funny. My error?
Error in (function (classes, fdef, mtable) :
unable to find an inherited method for function ‘dbWriteTable’ for signature ‘"ACCESS", "character", "function"’
@dmenne can you try this code to use db_write_table()? First load my Gist and my branch, then run the following:
# Use your connection here
con <- dbConnect(odbc::odbc(), dsn = "dbplyr-testing")
db_write_table(con = con, types = c(speed = "DOUBLE", dist = "DOUBLE"), table = "somecars", values = cars)
At the moment, you have to explicitly specify the types. This works for me.
The invalid precision error is one I have dealt with as well. Jim Hester and I believe the solution is actually NOT to create a new odbcDataType.ACCESS, as the table is actually created successfully:
> DBI::dbWriteTable(con, "somecars2", cars)
Error in result_insert_dataframe(rs@ptr, values) :
nanodbc/nanodbc.cpp:1791: HY104: [Microsoft][ODBC Microsoft Access Driver]Invalid precision value
> DBI::dbExistsTable(con, "somecars2")
[1] TRUE
> DBI::dbGetQuery(con, "SELECT * FROM somecars2")
[1] speed dist
<0 rows> (or 0-length row.names)
This is directly related to my open issue on odbc here.
"...the error occurs when binding parameters to the table." - Jim
I agree with @edgararuiz that this should be handled by odbc not dbplyr, and that people can begin using the read-only capabilities while writing tables is worked out.
I wonder how a table can be created in Access using the odbcDataType .default, it shouldn't work
Bingo ! That's the funny csv-workaround ...!
Strange that I have to explicitly give the filename in source_gist, but that's another issue.
library(odbc)
library(dplyr)
library(dbplyr)
library(DBI)
devtools::source_gist( "7d8dd9338afcd5c2eb94d751097f5296",
filename = "db_-access-implementation.R" )
cs = "Driver=Microsoft Access Driver (*.mdb, *.accdb);DBQ=./test.accdb"
con = dbConnect(odbc::odbc(), .connection_string = cs)
db_write_table(con = con, types = c(speed = "DOUBLE", dist = "DOUBLE"), table = "somecars", values = cars)
I have to correct my version 44 minutes ago: I had not noticed that library(rlang) was missing. Here again
#other attached packages:
# [1] DBI_0.7 dbplyr_1.1.0.9000 dplyr_0.7.2 odbc_1.1.1.9000
db_write_table(con = con, types = c(speed = "DOUBLE", dist = "DOUBLE"), table = "somecars", values = cars, append = TRUE, overwrite = FALSE)
Note: method with signature ‘DBIConnection#SQL’ chosen for function ‘dbQuoteIdentifier’, target signature ‘ACCESS#SQL’. "OdbcConnection#character" would also be valid Error: Unsupported type
@edgararuiz I think I know what's happening. I was using the CRAN version of odbc 1.1.1, and since then the defaults for odbcDataType have changed. The previous defaults just had double = "DOUBLE" which worked fine for table creation with Access.
https://github.com/rstats-db/odbc/commit/56fdd790a611808c0ef9d102caecd3ea93eb66cb
With the dev version of odbc I get the error and the table is not created! (because "DOUBLE PRECISION" does not exist as a type in Access)
> DBI::dbWriteTable(con, "somecars2", mtcars)
Error in new_result(connection@ptr, statement) :
nanodbc/nanodbc.cpp:1344: 42000: [Microsoft][ODBC Microsoft Access Driver] Syntax error in CREATE TABLE statement.
This does support your idea that we need a odbcDataType.ACCESS method
(As a side note, after installing dev odbc the Connections window started working and I am incredibly impressed)
A potential odbcDataType.ACCESS method.
Not yet fully vetted, but interestingly using logical = "BIT" transfers R logical vectors to and from Access correctly using my db_write_table(), so that's good news.
odbcDataType.ACCESS <- function(con, obj, ...) {
switch_type(obj,
factor = "TEXT",
datetime = "DATETIME",
date = "DATETIME",
time = "DATETIME",
binary = "BINARY",
integer = "INTEGER",
double = "DOUBLE",
character = "TEXT",
logical = "BIT", # only valid if DB supports Null fields
list = "INTEGER",
stop("Unsupported type", call. = FALSE)
)
}
@DavisVaughan - Are you saying that db_write_table() is now able to create new tables?
Yes, as far I can see, it's the way to go. The CSV-detour might play nasty tricks when dates/times are involved, I remember ugly debugging when I did the same by hand earlier. Do we have a test battery for these cases?
Ok, can you guys also try DBI::dbWriteTable() too please? If it works, we just need to send Jim a PR for the translation
If this is the correct syntax:
DBI::dbWriteTable(con, field.types = c(speed = "DOUBLE", dist = "DOUBLE"), append = TRUE)
Error in (function (classes, fdef, mtable) :
unable to find an inherited method for function ‘dbWriteTable’ for signature ‘"ACCESS", "missing", "missing"’
Cross-check: this works
db_write_table(con = con, types = c(speed = "DOUBLE", dist = "DOUBLE"), table = "somecars", values = cars)
DBI::dbWriteTable() does not work even with the odbcDataType.ACCESS fix. I just want to make sure we are on the same page here so:
dbWriteTable for odbc seems to be split into two main parts:
1) A part that creates an _empty_ table. This part works when we use odbcDataType.ACCESS().
2) A part that adds our data to that table using an Insert query and the C++ function result_insert_dataframe(). This is what fails with an invalid precision error. Jim thinks that somewhere inside that C++ function parameters are being bound incorrectly for Access SQL. For me this is really hard to debug, so I was waiting on Jim to look into it and come up with the solution. _If we can get him to do this, we are good to go._
In the mean time, I had created db_write_table.ACCESS to write to a text file and load that directly into Access instead of going through dbWriteTable and that odbc C++ function. While it works, I would much rather use the odbc implementation of dbWriteTable if we can get it working.
Just a quick ping to @edgararuiz, should I open a pull request for the read-me elements of this?
Yes, I think we should.
Most helpful comment
That's great!
If you plan to start soon, I'd suggest to use this Issue for communication. We can transition to something else if it becomes necessary.
To get you started, the translation should be a new R script in
dbplyrcalleddb-odbc-access.R. I'd suggest to start with a copy of the MSSQL translation (https://github.com/tidyverse/dbplyr/blob/master/R/db-odbc-mssql.R) and modify it to fit Access' syntax.