Data.table: data table gives error while trying to cut a numeric row based on some criteria
I have a data in SQLITE. I extract it just fine
sqlite <- dbConnect(SQLite(),
'www/main_data.sqlite')
fulldata<-sqlite %>%
tbl('employee_data') %>%
filter( between(date,'2018-01-01','2018-03-31')) %>%
filter(organization %in% c('nkid','honda')) %>%
collect()
fulldata %>% setDT()
the data looks like this
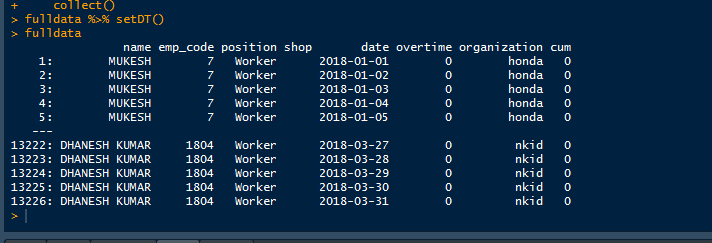
I calculate the running total of overtime on it. datewise
fulldata[,date:=as.Date(date)]
fulldata[is.na(overtime),overtime:=0]
setorder(fulldata,date)
fulldata[,cum:=cumsum(overtime),emp_code]
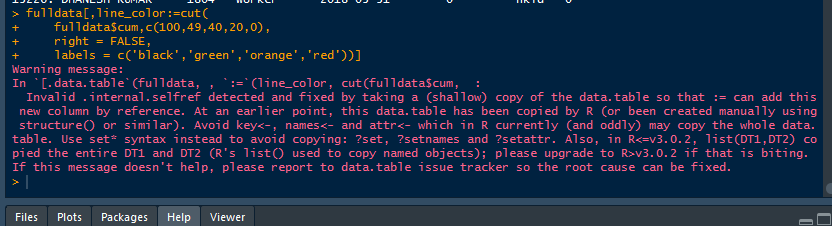
so far this is good there are no problems. but when I cut it into logical bins I get an error.
fulldata[,line_color:=cut(
fulldata$cum,c(100,49,40,20,0),
right = FALSE,
labels = c('black','green','orange','red'))]
this is the error
> fulldata[,line_color:=cut(
+ fulldata$cum,c(100,49,40,20,0),
+ right = FALSE,
+ labels = c('black','green','orange','red'))]
Warning message:
In `[.data.table`(fulldata, , `:=`(line_color, cut(fulldata$cum, :
Invalid .internal.selfref detected and fixed by taking a (shallow) copy of the data.table so that := can add this new column by reference. At an earlier point, this data.table has been copied by R (or been created manually using structure() or similar). Avoid key<-, names<- and attr<- which in R currently (and oddly) may copy the whole data.table. Use set* syntax instead to avoid copying: ?set, ?setnames and ?setattr. Also, in R<=v3.0.2, list(DT1,DT2) copied the entire DT1 and DT2 (R's list() used to copy named objects); please upgrade to R>v3.0.2 if that is biting. If this message doesn't help, please report to data.table issue tracker so the root cause can be fixed.
>
I am running R 3.5 and the data.table package is updated too.
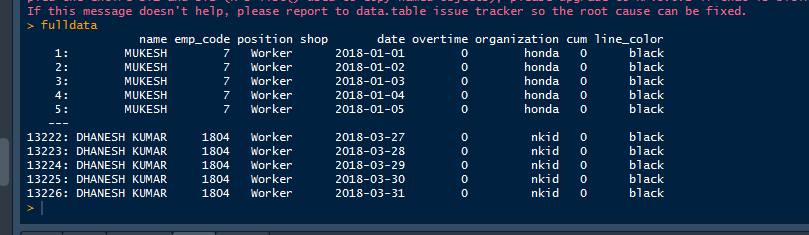
the funny part is that it does create the column for me but still gives me an error. the new Datatable looks like this.
But because of this error I am not able to run it in a shiny application. Where I need it.
Please let me know what's wrong and if there is anything I can do about it.
All 10 comments
Why aren't the breaks sorted? I guess they don't need to be (cut(runif(100), c(.5, 0, 1)) runs fine).
Error is strange anyway. Can you share fulldata?
I was asking for help in community.rstudio.com so I have already established a database in
Cloud.rstudio.com
Just click on this link
https://rstudio.cloud/project/36144
And it will take you on the project
The data is in the www folder. You can take a look at that.
And thanks for such a prompt reply.
this is being deployed on a shiny? could you include the "server" part of
your code as well?
On Sat, May 12, 2018, 2:14 PM Anantadinath notifications@github.com wrote:
I was asking for help in community.rstudio.com so I have already
established a database in
Cloud.rstudio.comJust click on this link
https://rstudio.cloud/project/36144 3
And it will take you on the project—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub
https://github.com/Rdatatable/data.table/issues/2869#issuecomment-388533313,
or mute the thread
https://github.com/notifications/unsubscribe-auth/AHQQdThd4FlRlTXof9yYy3mPnLldz6rdks5txn21gaJpZM4T8SqT
.
I guess you don't need that.
Cloud.rstudio.com provides an r session on cloud.
It has a database in www folder and the code that I have written on the top are there to extract it and modify it.
And just for information three server code is also written in the code as it is a single file app.
Please I understand you would have priorities but if you can just click on the link and run the codes there. It's just that I am not in a situation to upload any data right now. I am outside in a clients location.
Please if you could just click on the link. You will get an extract replica of my data..
My point is it should not produce an error on this operation
not looking for the data here, but the exact code you're running. there are
some other outstanding issues where that .internal.selfref error can arise,
and seeing that code can help contextualize your error vs. the other
related errors (the root cause might be the same, for example)
On Sat, May 12, 2018, 2:46 PM Anantadinath notifications@github.com wrote:
I guess you don't need that.
Cloud.rstudio.com provides an r session on cloud.
It has a database in www folder and the code that I have written on the
top are there to extract it and modify it.And just for information three server code is also written in the code as
it is a single file app.Please I understand you would have priorities but if you can just click on
the link and run the codes there. It's just that I am not in a situation to
upload any data right now. I am outside in a clients location.Please if you could just click on the link. You will get an extract
replica of my data..—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub
https://github.com/Rdatatable/data.table/issues/2869#issuecomment-388534826,
or mute the thread
https://github.com/notifications/unsubscribe-auth/AHQQdXxbMLKFBRBs4U9itFXM7TNl3e-zks5txoU2gaJpZM4T8SqT
.
as a workaround if you're pressed for time, it looks like your data is
pretty small. you should be able to run copy() on your data before adding
the cut() column without much performance cost. That should make the
.internal.selfref error go away
On Sat, May 12, 2018, 3:03 PM Michael Chirico michaelchirico4@gmail.com
wrote:
not looking for the data here, but the exact code you're running. there
are some other outstanding issues where that .internal.selfref error can
arise, and seeing that code can help contextualize your error vs. the other
related errors (the root cause might be the same, for example)On Sat, May 12, 2018, 2:46 PM Anantadinath notifications@github.com
wrote:I guess you don't need that.
Cloud.rstudio.com provides an r session on cloud.
It has a database in www folder and the code that I have written on the
top are there to extract it and modify it.And just for information three server code is also written in the code as
it is a single file app.Please I understand you would have priorities but if you can just click
on the link and run the codes there. It's just that I am not in a situation
to upload any data right now. I am outside in a clients location.Please if you could just click on the link. You will get an extract
replica of my data..—
You are receiving this because you commented.
Reply to this email directly, view it on GitHub
https://github.com/Rdatatable/data.table/issues/2869#issuecomment-388534826,
or mute the thread
https://github.com/notifications/unsubscribe-auth/AHQQdXxbMLKFBRBs4U9itFXM7TNl3e-zks5txoU2gaJpZM4T8SqT
.
I got it now. Thanks for replying again. I am not pressed in time. I am just not in front of a system here. I will sure give you exact code that causes problem. One I reach home. Thanks again..
I understood what you said and I ran the same code on the cloud.RStudio.com. Just to confirm that I am not facing this issue on my own computer.
this is the screenshot it gave me the same exact error.
I even tried using an if-else approach to data.table instead of cut. but it still gives me the same error. I have pasted all the code in cloud.Rstudio.com check them at your time. and Thanks for helping me up.
fulldata[,line_color:=ifelse(
cum<20,'black',ifelse(cum>=20 & cum <40,'green',
ifelse(cum>=40 & cum<49,'orange',
'red')))]
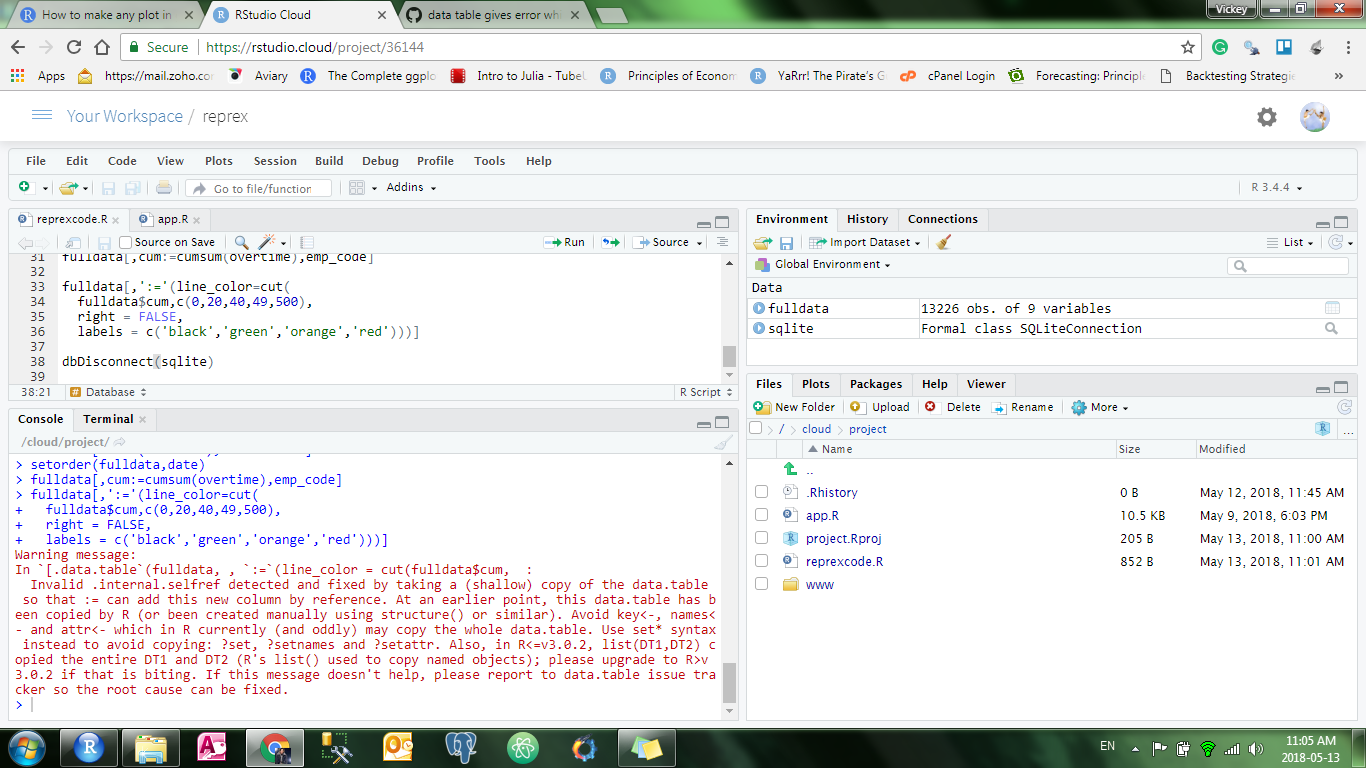
and this is the sort of error it generates when I try to run shiny application. Because it manipulates the data everytime I filter it.
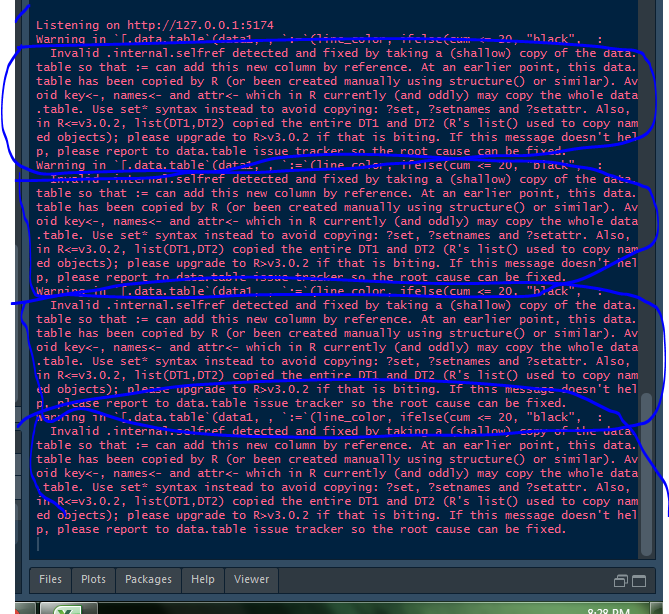
> sessionInfo()
R version 3.5.0 (2018-04-23)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 7 x64 (build 7601) Service Pack 1
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DT_0.4 crosstalk_1.0.0 DBI_1.0.0
[4] magrittr_1.5 dplyr_0.7.4 dbplyr_1.2.1
[7] lubridate_1.7.4 data.table_1.11.2 plotly_4.7.1
[10] ggplot2_2.2.1.9000 shinydashboard_0.7.0 shiny_1.0.5
[13] RSQLite_2.1.1
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 compiler_3.5.0 pillar_1.2.2 later_0.7.2
[5] plyr_1.8.4 bindr_0.1.1 tools_3.5.0 digest_0.6.15
[9] bit_1.1-12 viridisLite_0.3.0 jsonlite_1.5 memoise_1.1.0
[13] tibble_1.4.2 gtable_0.2.0 pkgconfig_2.0.1 rlang_0.2.0
[17] yaml_2.1.19 bindrcpp_0.2.2 stringr_1.3.1 httr_1.3.1
[21] withr_2.1.2 htmlwidgets_1.2 bit64_0.9-7 grid_3.5.0
[25] glue_1.2.0 R6_2.2.2 tidyr_0.8.0 purrr_0.2.4
[29] blob_1.1.1 scales_0.5.0 promises_1.0.1 htmltools_0.3.6
[33] rsconnect_0.8.8 assertthat_0.2.0 mime_0.5 xtable_1.8-2
[37] colorspace_1.3-2 httpuv_1.4.3 stringi_1.2.2 lazyeval_0.2.1
[41] munsell_0.4.3
Thank you, and duly noted for when this issue will be addressed 😃