Data.table: fread spends too much time in is_url/is_secureurl/is_file for long in memory input
Summary
When fread is fed with a character string as input the routine spends considerable amount of time detecting that the supplied input is not a filename or an url.
This is due to grepl not scaling well for large input as used in fread.
Example
is_url <- function(x) grepl("^(http|ftp)s?://", x)
Although the pattern is anchored at the beginning of the string, running grepl for large inputs will take a lot of time for large inputs (more detailed benchmarks further down).
This will lead then to the full call to fread possibly spending a third of the time for those supposedly simple checks. (example also below)
Possible solutions could be one or more of the following
- changing the call to
greplto use PERL regexp engineperl=TRUE
Alternatively use another method to determine whether the input starts with url (see benchmarks below) - adding an explicit optional argument which denotes string input, e.g.
str=to denote that the input is to be considered as the data and skip the tests for url or file. This would be similar in spirit to thefile=argument.
As a side-effect this would also allow input to be read which consits only of url's and having no header, e.g.
```R
fread("http://hkhfsk\nhttp://fhdkf\nhttp://kjfhskd\nhttp://hfkjf", header=FALSE)
* consider an input, which exceeds a certain number of characters in length, not being an URL
## Profiling example
```R
library(data.table)
# create a random character input in 5 columns with 20000*200 lines
randomString <- function() {
paste(sample(c(LETTERS, letters), sample(1:9,1)), collapse="")
}
input <- paste(rep(paste(replicate(20000,paste(replicate(5, randomString()),
collapse="\t")),
collapse="\n"),200),
collapse="\n")
# profile the call to fread
Rprof()
invisible(fread(input, header=FALSE))
Rprof(NULL)
summaryRprof()
```
$by.self
self.time self.pct total.time total.pct
".Call" 5.34 64.49 5.34 64.49
"grepl" 2.94 35.51 2.94 35.51
## Benchmarking `grepl` and friends
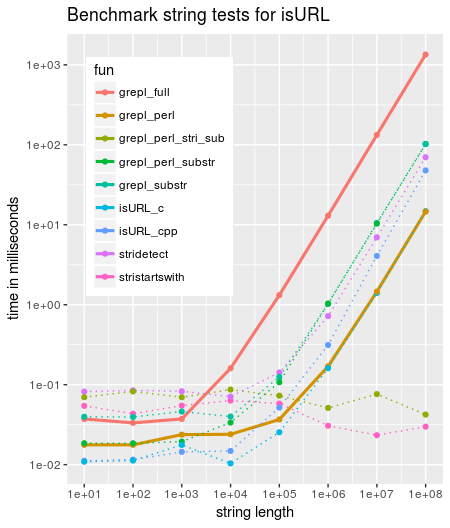
Comparing different functions to verify whether a string starts with any of http(s)/ftp(s) or file shows that `grepl` scales badly and is by far the slowest of the tested variants.
Adding simply `perl=TRUE` already improves by around factor 100 for large inputs (code further below)
```R
library(data.table)
library(ggplot2)
library(Rcpp)
# define alternative functions to detect URL's
grepl_full <- function(txt) { grepl("^(http|ftp)s?://", txt) }
grepl_substr <- function(txt) { grepl("^(http|ftp)s?://", substr(txt,1,8)) }
grepl_perl <- function(txt) { grepl("^(http|ftp)s?://", txt, perl=TRUE) }
grepl_perl_substr <- function(txt) { grepl("^(http|ftp)s?://", substr(txt,1,8), perl=TRUE) }
grepl_perl_stri_sub <- function(txt) { grepl("^(http|ftp)s?://", stringi::stri_sub(txt,1,8),
perl=TRUE) }
stridetect <- function(txt) { stringi::stri_detect_regex(txt, "^(http|ftp)s?://") }
stristartswith <- function(txt) {
stringi::stri_startswith_fixed(txt, c("http://", "https://", "ftp://", "ftps://"))
}
cppFunction('bool isURL_cpp(const std::string &x) {
bool ret = FALSE;
if (x.length() >= 8) {
std::string s = x.substr(0,8);
if (s.substr(0,7) == "http://" ||
s == "https://" ||
s.substr(0,6) == "ftp://" ||
s.substr(0,7) == "ftps://") {
ret = TRUE;
}
}
return(ret);
}')
cppFunction('bool isURL_c(const char* x) {
bool ret = FALSE;
if (strlen(x) >= 8) {
if (!strncmp(x, "http://", 7) ||
!strncmp(x, "https://", 8) ||
!strncmp(x, "ftp://", 6) ||
!strncmp(x, "ftps://", 7)) {
ret = TRUE;
}
}
return(ret);
}')
# number of string lengths to test with. max len = 1e+nn
nn <- 8
# create random strings of given length
for (i in 1:nn) {
assign(paste0("str_",i), paste(sample(c(LETTERS, letters), 10^i, replace=TRUE),
collapse=""))
}
function_list <- c("grepl_full", "grepl_substr", "grepl_perl", "grepl_perl_substr",
"grepl_perl_stri_sub",
"isURL_cpp", "isURL_c",
"stridetect", "stristartswith")
# cross join of functions and strings
dt <- CJ(fun=function_list, ind=1:nn)
# build code to run microbenchmark
mb_code <- paste("microbenchmark::microbenchmark(",
paste0("'", dt$fun, " ", 10^dt$ind, "'=",
dt$fun, "(str_", dt$ind, ")", collapse=","),",times=10)")
# eval microbenchmark and calculate median
mb <- as.data.table(eval(parse(text=mb_code)))
mb[,c("fun", "nr"):=tstrsplit(as.character(expr), " ", fixed=TRUE)]
mb[,nr:=as.integer(nr)]
mb <- mb[,.(time=median(time)), by=.(fun,nr)]
# plot
ggplot(mapping=aes(nr, time/1e6)) +
geom_line(data=mb[fun %chin% c("grepl_full", "grepl_perl")],
aes(col=fun), size=1.1) +
geom_line(data=mb[!fun %chin% c("grepl_full", "grepl_perl")],
aes(col=fun), size=0.5, linetype="dotted") +
geom_point(data=mb, aes(col=fun)) +
scale_y_log10(breaks=c(0.01,0.1,1,10,100,1000)) +
scale_x_log10(breaks=10^(1:nn)) +
labs(y="time in milliseconds", x="string length",
title="Benchmark string tests for isURL") +
theme(legend.position = c(0.05,0.95), legend.justification = c(0,1))
sessionInfo
R version 3.3.1 (2016-06-21)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.3 LTS
locale:
[1] LC_CTYPE=en_GB.UTF-8 LC_NUMERIC=C LC_TIME=en_IE.UTF-8 LC_COLLATE=en_GB.UTF-8
[5] LC_MONETARY=en_IE.UTF-8 LC_MESSAGES=en_GB.UTF-8 LC_PAPER=en_IE.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_IE.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.10.5
loaded via a namespace (and not attached):
[1] tools_3.3.1 yaml_2.1.15
Results are similar with R 3.4.3 / data.table 1.10.4 / Windows 10 64bit
All 9 comments
Seems like a problem of the regex engine more than anything. anchoring to string beginning should mean that grepl performance is O(1), as far as I'm concerned. Strange that it appears otherwise.
probably R's regex engine attempts to build a histogram of the target string before applying the regexp.
i have in mind this:
Indeed it may be more a R issue, but something one can at least work around in fread
Using perl=TRUE does not seem to add any negative aspect and will be usally faster (in my benchmarks 100 times for large input)
See also help-page: ?grep:
If you are doing a lot of regular expression matching, including on very long strings, you will want to consider the options used. Generally PCRE will be faster than the default regular expression engine,
The additional penalty (even when using perl=TRUE) does not seem to come from grep: Look at the benchmark when feeding grep with the shortened string though substr (grep_perl_substr) whereas using stringi::stri_sub performs with O(1) (grep_perl_strisub). So there seems to be an overhead when calling the base function with a large string. (Actually we see this even for the Rcpp versions)
For later reference, the utility startsWith is easily the fastest (and clearest), but unfortunately depends on R (>= 3.3.0).
is_url <- function(x) grepl("^(http|ftp)s?://", x)
is_url_perl <- function(x) grepl("^(?:http|ftp)s?://", x, perl = TRUE)
is_url2 <- function(x) {
`||`(startsWith(x, "ht") && startsWith(x, "http://") || startsWith(x, "https://"),
startsWith(x, "ft") && startsWith(x, "ftp://") || startsWith(x, "ftps://"))
}
microbenchmark::microbenchmark(is_url(input), is_url_perl(input), is_url2(input), times = 50)
Unit: microseconds
# expr min lq mean median uq max neval cld
# is_url(input) 1001673.642 1038144.688 1076978.567 1070702.5405 1112537.553 1185685.65 50 c
# is_url_perl(input) 23280.392 24793.807 30226.567 27345.3795 30737.237 51998.24 50 b
# is_url2(input) 1.807 2.711 139.927 12.8005 18.975 6414.17 50 a
URL <- "https://github.com/Rdatatable/data.table/issues/2531"
microbenchmark::microbenchmark(is_url(URL), is_url_perl(URL), is_url2(URL), times = 50)
# expr min lq mean median uq max neval cld
# is_url(URL) 6.626 7.228 8.40290 7.680 8.132 30.118 50 b
# is_url_perl(URL) 65.657 66.259 67.40972 66.560 66.862 92.462 50 c
# is_url2(URL) 1.506 1.808 2.25304 2.108 2.409 8.132 50 a
This was a great report -- many thanks!
I've just fixed it and pushed to master. Please test and reopen if this doesn't fix it. I'm pretty sure it's much better now, but you never know. I pushed it straight to master (rather than a PR) so you can more easily test it just by upgrading to latest dev.
I added your test too so it now reads those http:// addresses as data as you suggested.
Thank you for looking into this issue.
I am afraid it is not fixing the issue of slow execution time completely.
In the new code, once you go in the else clause of if (!missing(file)) the first thing you are checking (after verifying input is a length 1 character vector) is
if (file.exists(input)) {
....
unfortunately file.exists is also very slow for large input:
input <- paste(rep("1,2,3,4.567,some text field", 1e6),collapse="\n")
system.time(file.exists(input))
## user system elapsed
## 0.693 0.000 0.694
system.time(fread(input))
## user system elapsed
## 0.861 0.000 0.862
with the file.exists eating up 80% of the total fread execution time.
Fortunately the next check you are doing with grep (detecting newlines) is very fast (opposed to the suprisingly slow grepl checks:
system.time(length(grep('\\n|\\r', input)))
## user system elapsed
## 0 0 0
The easiest fix seems to be to swap the two conditions. The only case where I see this would make a difference is, if input is a filename and has a newline in the name, but I hope no ones would do this and then expect fread to be able to read the file. (and actually it wouldn't work as the c-routine for fread checks again for newlines and would treat such a file name as the actual input)
You suggested me to re-open the issue in case it's not working, but as reporter, I can not re-open it.
I have opened a PR #2630 with the proposed fix.
The other part of your fix with substring etc is indeed avoiding grepl is slightly faster, but due to the fact you check now for newlines earlier in the code the penalty should not be visible ... unless someone is giving as input a very long string _without_ any newlines ... but that seems also like a pathological case.
Thanks for the follow up. Yes I had thought file.exists() would be fast on huge input as the operating system would detect it was invalid in the first few characters. Interesting it isn't. Maybe R passes over it first before calling the OS for some reason. Anyway, will proceed with your PR, thanks.
Didn't know you can't reopen issues; must be just project members that see that option then. I'll see if there's a project setting to open that, and will bear it in mind in future. I've invited you to be project member too.
just for the record to fully close it.
With the changes applied in 8ab3028d867ac29a9cca1d2b709a0758ce74ee9c and 3989ea20f000c2ae95d7301e97e467be222c4992 fread speedup for data input through the input variable is substantial.
For input with > 0.5 MB size / 10000 rows the speedup between before and after fixes is 3 - 5.

Most helpful comment
just for the record to fully close it.
With the changes applied in 8ab3028d867ac29a9cca1d2b709a0758ce74ee9c and 3989ea20f000c2ae95d7301e97e467be222c4992
freadspeedup for data input through the input variable is substantial.For input with > 0.5 MB size / 10000 rows the speedup between before and after fixes is 3 - 5.