Chapel: Support Jupyter Kernel for Chapel
In doing iterative research, being able to experiment and log results is invaluable. One extremely popular tool for this is Jupyter
The Chapel IPE and Chapel TIO suggest Chapel could be run within Jupyter. This would be awesome.
Please create a Jupyter kernel for Chapel.
All 18 comments
Until we have a true interpreter or JIT, the approach taken with the jupyter-c-kernel seems the best for now.
I was going to make a comment almost identical to Ben's, but he beat me to it. I've never looked into the Jupyter C kernel -- can someone summarize the approach it takes and how it works around the JIT issue for C?
how it works around the JIT issue for C?
It doesn't - see: https://github.com/brendan-rius/jupyter-c-kernel/issues/28
The relationship between multiple cells and JITing isn't clear to me. Is the implication that they recompile the entire program anytime it's edited?
I haven't used it myself, but that seems to be the case, at best.
The Jupyter C kernel indeed recompiles every cell, I'm looking for a good way of splitting the implementation over multiple cells. There is a related open Stack Overflow question on this as well (SO-43316613) from the maintainer of the jupyter-elm-kernel.
He implemented a workaround by adding an explicit -- compile-code flag.
If you ever find out a nice/clean way, I would love to hear it.
Thanks for the information and pointers @spoorcc!
A user has developed some early chapel magic function support here
@ben-albrecht @bradcray @lydia-duncan Is it possible to take this as a GSOC project?
@krishnadey30 - I think this could make a good GSoC project, if carefully scoped. For example, we may want to pursue the recompile-entire-notebook approach as a first step, and then investigate some of the improvements discussed above.
@ben-albrecht I will try to come up with a good proposal for this project and will consider the improvements discussed above.
@buddha314 @ben-albrecht @bradcray You might want to look at this.
Xeus-cling is a Jupyter kernel for C++ based on the C++ interpreter cling and the native implementation of the Jupyter protocol xeus.
@krishnadey30 - looks like the link above is broken, here's another link: https://github.com/QuantStack/xeus-cling
Interesting project. Do you know if they evaluate cell-by-cell rather than recompiling/running entire notebook per evaluation? If so, do you know how they are implementing this?
@ben-albrecht I have updated the link. I think they evaluate cell by cell but I need to check it more properly. I am going through the documentation of Xeus and Xeus-Cling. I will look at Cling and let you know.
@krishnadey30 - looks like the link above is broken, here's another link: https://github.com/QuantStack/xeus-cling
Interesting project. Do you know if they evaluate cell-by-cell rather than recompiling/running entire notebook per evaluation? If so, do you know how they are implementing this?
Cern states that Cling is an interactive C++ interpreter and uses just-in-time (JIT) compiler for compilation.
Also, I tried the online notebook that has been provided by them and found this. This might give the answer.
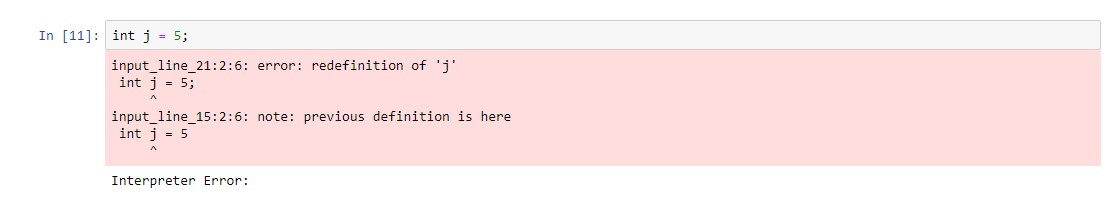
I have created a python wrapper kernel for Chapel. https://github.com/krishnadey30/jupyter_kernel_chapel
With the creation of jupyter-kernel-chapel and the ability to install it via pip (pip3 install jupyter-kernel-chapel), I'm inclined to close this issue and redirect further Jupyter kernel improvement requests to that repository.
Would anyone oppose closing this issue?
Most helpful comment
I have created a python wrapper kernel for Chapel. https://github.com/krishnadey30/jupyter_kernel_chapel